Comprehensive Molecular Profiling Matches Pediatric Patients to Therapies
By LabMedica International staff writers
Posted on 22 Oct 2020
A major challenge of using a comprehensive profiling approach for precision medicine is separating pathogenic from non-pathogenic molecular changes within a tumor.Posted on 22 Oct 2020
A program has been presented of the first systematic evaluation of the utility and early clinical effects of a comprehensive molecular profiling platform to identify clinically significant variants relevant to the biology of the tumor, diagnosis, clinical management, or prognosis.
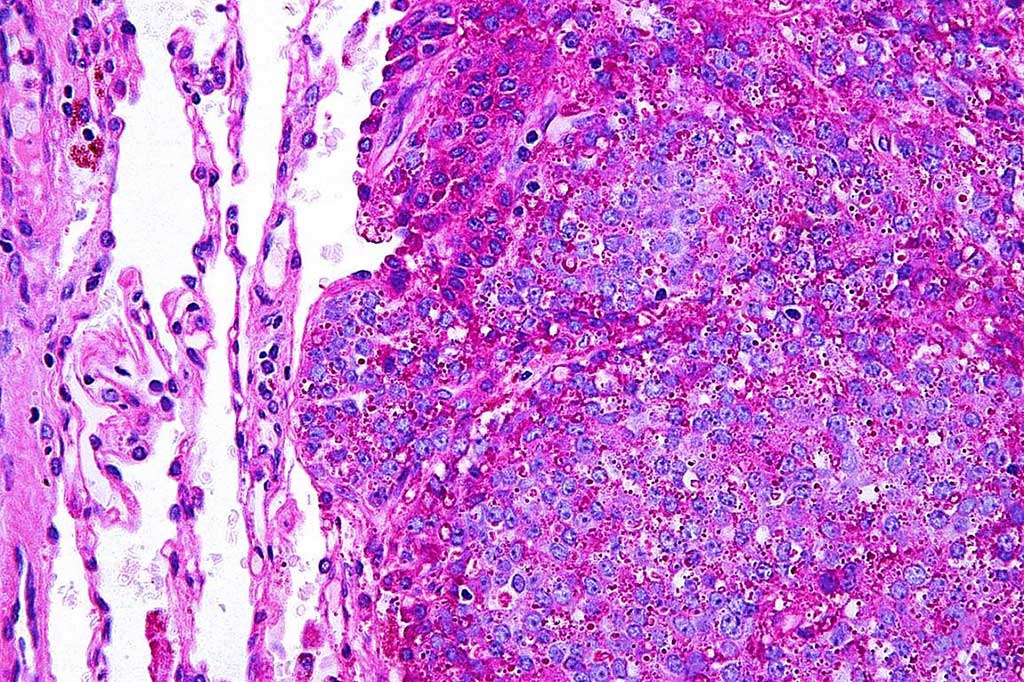
Image: Photomicrograph of histology of a Ewing sarcoma in lung. The PAS stain highlights glycogen and the staining was negative with a PAS diastase stain (Photo courtesy of Nephron).
Oncologists at the Children’s Cancer Institute (Kensington, Australia) analyzed 252 tumors from high-risk pediatric patients with cancer. These patients typically have aggressive tumors, but have few treatment options, or relapsed or refractory disease despite having undergone standard therapy. The team used tumor and germline whole genome sequencing (WGS) and RNA sequencing (RNAseq) across all the tumors from these high-risk pediatric patients with cancer.
The analysis identified 968 reportable molecular aberrations, 39.9% through WGS and RNA-seq, 35.1% through WGS only, and 25% through RNA-seq. Of these patients, 93.7% had at least one germline or somatic aberration, 71.4% had molecular aberrations that could be treated with a targeted therapy, and 5.2% had a change in diagnosis. The investigators also noted that WGS identified pathogenic cancer-predisposing variants in 16.2% of the patients. When they conducted methylome analysis in 76 central nervous system tumors, diagnosis was confirmed in 71.1% of patients and contributed to a change of diagnosis in two patients.
The scientists sought to identify non-coding driver variants linked to unexplained gene expression changes, and identified 67 established or novel driver fusions using integrated WGS and RNA-seq. The most frequent were EWSR1 rearrangements, PAX3-FOXO1, and ASPCR1-TFE3 in Ewing's sarcoma, alveolar rhabdomyosarcoma, and alveolar soft part sarcoma, respectively. They noted that there were 15 likely kinase-activating fusions, including six NTRK fusions, and that other fusions highlighted new tumor biology.
When the investigators looked for therapeutically actionable single-nucleotide variants (SNVs), copy number variants (CNVs), and structural variants, they found them clustered most frequently in RTK signaling, MAP kinase signaling, and PI3K-mTOR signaling pathways. RTKs were activated by point mutations, high copy number gains, or fusions. PI3K-mTOR variants included known activating PIK3CA mutations or loss-of-function mutations and deletions affecting PTEN, PTPN11, PIK3R1, PIK3R2, MTORC1, and MTORC2. The team also recurrently observed potentially targetable variants in epigenetic regulation genes and in the chromatin-remodeling gene PBRM1.
The authors concluded that this was the first estimate of the prevalence of germline mutations driving high-risk pediatric cancer in the Australian population based on tumor-germline WGS. WGS was critical for the assignment of pathogenicity to several variants, notably where somatic features, such as mutation signatures, tumor mutational burden (TMB) and second-hit mutations, were instrumental in identifying the underlying germline mutation(s). The study was published on October 5, 2020 in the journal Nature Medicine.
Related Links:
Children’s Cancer Institute