Rapid CRISPR/Cas Assay for Diagnosis of Viral Fevers in Low-Tech Environments
By LabMedica International staff writers
Posted on 07 Sep 2020
Rapid diagnostic tests for Ebola and Lassa fever viruses based on the RNA-targeting CRISPR/Cas13a complex have been tested successfully under field conditions in several African countries.Posted on 07 Sep 2020
COVID-19 is not the only disease out there. Recent outbreaks of viral hemorrhagic fevers (VHFs), including Ebola virus disease (EVD) and Lassa fever (LF), highlight the urgent need for sensitive, deployable tests to diagnose these devastating human diseases. In this regard, investigators at the Broad Institute of MIT and Harvard (Cambridge, MA, USA) and their collaborators developed CRISPR-Cas13a-based diagnostics targeting Ebola virus (EBOV) and Lassa virus (LASV), with both fluorescent and lateral flow readouts.
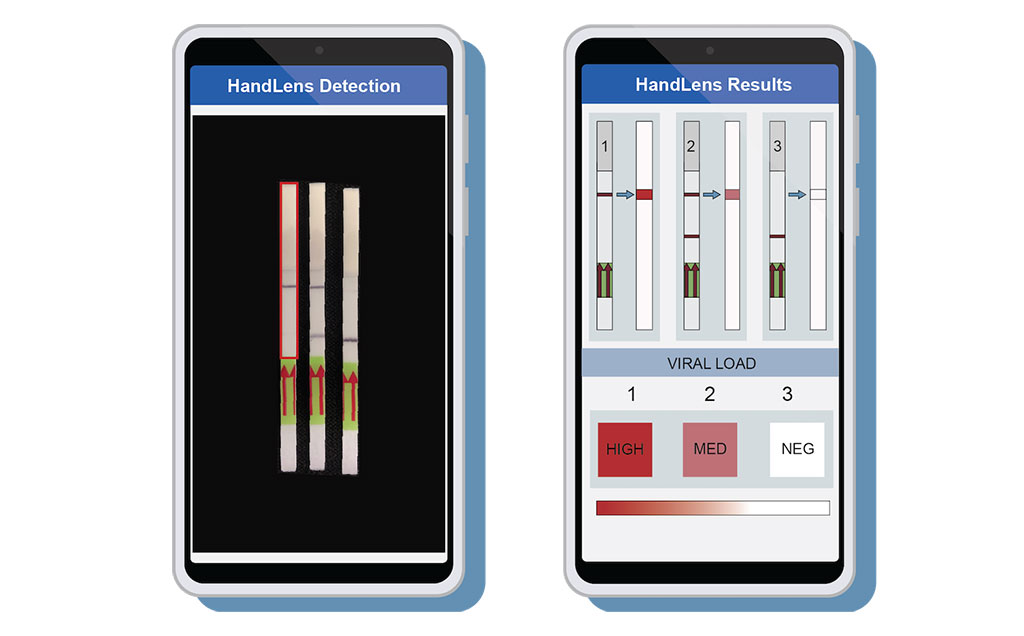
Image: A mobile phone app called HandLens was designed to read and report paper strip SHERLOCK results (Photo courtesy of Anna Lachenauer, Stanford University School of Medicine)
CRISPRs (clustered regularly interspaced short palindromic repeats) are segments of prokaryotic DNA containing short repetitions of base sequences. Each repetition is followed by short segments of "spacer DNA" from previous exposures to a bacterial virus or plasmid. Since 2013, the CRISPR/Cas9 system has been used in research for gene editing (adding, disrupting, or changing the sequence of specific genes) and gene regulation. By delivering the Cas9 enzyme and appropriate guide RNAs (sgRNAs) into a cell, the organism's genome can be cut at any desired location.
Recent computational efforts to identify new CRISPR systems uncovered a novel type of RNA targeting enzyme, Cas13. The diverse Cas13 family contains at least four known subtypes, including Cas13a (formerly C2c2), Cas13b, Cas13c, and Cas13d. Cas13a was shown to bind and cleave RNA, protecting bacteria from RNA phages and serving as a powerful platform for RNA manipulation. It was suggested that Cas13a could function as part of a versatile, RNA-guided RNA-targeting CRISPR/Cas system holding great potential for precise, robust, and scalable RNA-guided RNA-targeting applications.
In developing an approach for rapidly adaptable, deployable diagnostics, the investigators worked with the CRISPR-based SHERLOCK (Specific High-sensitivity Enzymatic Reporter unLOCKing) platform. SHERLOCK utilizes the RNA-targeting protein Cas13a for sensitive and specific detection of viral nucleic acid. This is method works by amplifying genetic sequences and programming a CRISPR molecule to detect the presence of a specific genetic signature in a sample, which can also be quantified. When it finds those signatures, the CRISPR enzyme is activated and releases a robust signal. This signal can be adapted to work on a simple paper strip test, in laboratory equipment, or to provide an electrochemical readout that can be read with a mobile phone.
SHERLOCK can be combined with the HUDSON (Heating Unextracted Diagnostic Samples to Obliterate Nucleases) method, which inactivates pathogens and releases nucleic acid through a combined heat and chemical denaturation, eliminating the need for a column- or bead-based nucleic acid extraction. This process renders the patient samples safer for clinical staff to handle in low-tech environments, and eliminates the need to extract a virus's genetic material from the samples before analysis.
To complete the package, the investigators also developed a mobile phone application called HandLens, which can read and immediately report paper strip SHERLOCK results. This tool can aid in situations where the paper strip gives a faint signal that is challenging for a clinician to interpret.
The investigators used the SHERLOCK technique in conjunction with HUDSON and HandLens in Nigeria during a recent Lassa fever outbreak and in Sierra Leone and the Democratic Republic of the Congo during outbreaks of Ebola virus. Results revealed that the SHERLOCK assays performed as consistently as, or better than, other diagnostic methodologies, demonstrating the platform's potential for clinical use in resource-limited areas.
The SHERLOCK technique for rapid diagnosis of viral diseases was described in the August 17, 2020, online edition of the journal Nature Communications.
Related Links:
Broad Institute of MIT and Harvard














