CtDNA Assay for Cancer Recurrence, MRD Applications Demonstrated
By LabMedica International staff writers
Posted on 22 Jul 2020
Circulating tumor DNA (ctDNA) is found in the bloodstream and refers to DNA that comes from cancerous cells and tumors. Most DNA is inside a cell's nucleus. As a tumor grows, cells die and are replaced by new ones. The dead cells get broken down and their contents, including DNA, are released into the bloodstream.Posted on 22 Jul 2020
Cancer recurrence rates vary widely between cancer types, and within cancer types according to stage, histology, genetic factors, patient-related factors, and treatments. Minimal residual disease (MRD) is the name given to small numbers of leukemic cells (cancer cells from the bone marrow) that remain in the person during treatment or after treatment when the patient is in remission (no symptoms or signs of disease). It is the major cause of relapse in cancer and leukemia.
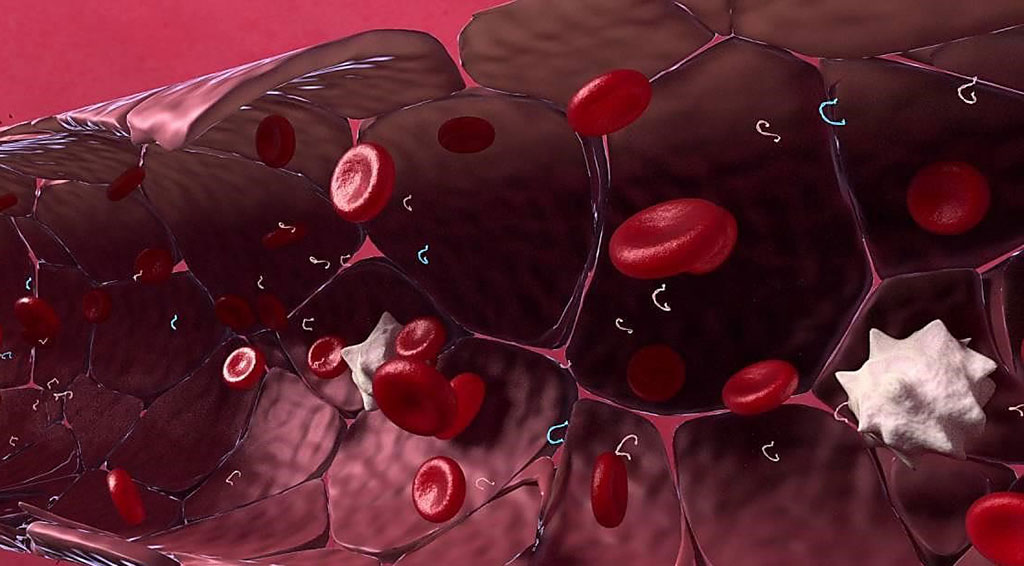
Image: The RaDaR (Residual Disease and Recurrence) assay detects circulating tumor DNA (ctDNA) found in the bloodstream (Photo courtesy of Inivata).
Scientists from Cancer Research UK Cambridge Institute (Cambridge, UK) analyzed three cancer cell lines at different dilutions, DNA reference material, seven formalin-fixed paraffin-embedded samples (from breast, colon, and melanoma), as well as 366 lung cancer samples from the Lung cancer-CIrculating tumor DNA (LUCID) study. They used DNA inputs of 20,000 or 4,000 copies.
The team used the RaDaR (Residual Disease and Recurrence) assay (Inivata, Babraham, UK). Inivata's RaDaR patient-specific assay runs on the company's InVision sequencing platform, which is built on a technology called targeted amplicon sequencing. RaDaR profiles a patient's tumor from a resection surgery and can target up to 48 unique mutations in the patient's cancer. After performing whole-exome sequencing (WES) on a patient's tumor tissue to identify somatic variants, Inivata ranks and prioritizes the variants based on proprietary algorithms to design a patient-specific panel.
The investigators using 48 mutations found that RaDaR had a sensitivity of 97% using 20,000 copies, and of 63% using 4,000 copies at a variant dilution of 20 parts per million (equivalent to a variant allele frequency of 0.002%), with 100% specificity. When looking at a subset of 16 mutations, RaDaR produced a sensitivity of 97% at 40 parts per million, 75% at 20 parts per million, and 38% at 10 parts per million, also with 100% specificity. The team’s colleagues carried out a second study and evaluated RaDaR's prognostic value and its ability to detect ctDNA prior to or during relapse in stage I to stage III non-small cell lung cancer (NSCLC) patients treated with curative intent with similar success.
Karen Howarth, DPhil, director of cancer genomics at Inivata and co-author of the studies said, “If you're putting 10,000 copies into a reaction, and you're looking for one mutant molecule, you're looking at one molecule in a background of 10,000. But, if you split it into four samples using partitioning, you're only looking at one molecule in 2,500 molecules, which pushes up the signal. The studies were presented at the 2020 American Association for Cancer Research Virtual Meeting held June 22 - 24, 2020.
Related Links:
Cancer Research UK Cambridge Institute
Inivata