Highly Sensitive Liquid Biopsy Technique Brings Personalized Cancer Monitoring a Step Closer
By LabMedica International staff writers
Posted on 29 Jun 2020
A modified liquid biopsy method enables detection of circulating tumor DNA (ctDNA) with much greater sensitivity than previously used techniques and demonstrates the potential to revolutionize all aspects of cancer care, from early detection to personalized treatment and monitoring.Posted on 29 Jun 2020
While ctDNA can be used to monitor cancer dynamics noninvasively, detecting it can be challenging in patients with low-volume or residual disease, where the plasma contains very few tumor-derived DNA fragments. Investigators at the University of Cambridge (United Kingdom) and their collaborators sought to develop a method for ctDNA detection with greatly increased sensitivity. To this end, they showed that sensitivity for ctDNA detection in plasma could be improved by using the INtegration of VAriant Reads (INVAR) pipeline. This technique combined custom error-suppression methods and signal-enrichment approaches based on biological features of ctDNA to analyze hundreds to thousands of mutations.
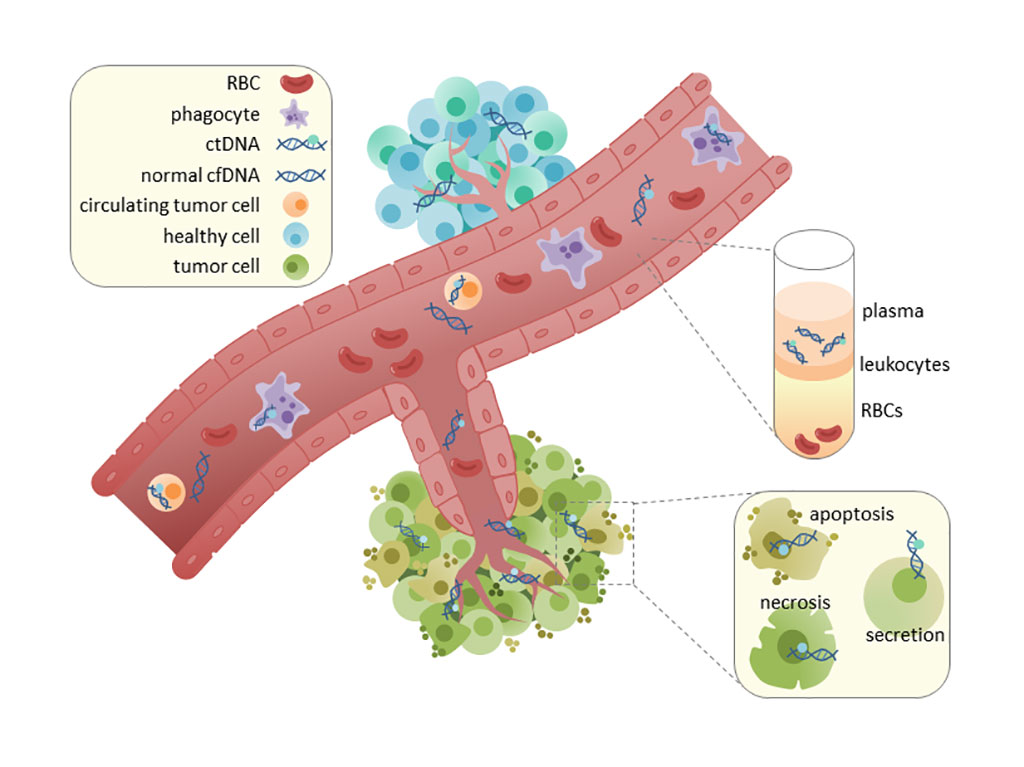
Image: Circulating tumor DNA (ctDNA) is found in serum and plasma fractions from blood. The mechanism of ctDNA release is unknown, though apoptosis, necrosis, and active secretion from tumor cells have been hypothesized. Once ctDNA is isolated, it can be sequenced for mutational analysis (Photo courtesy of Wikimedia Commons)
Current liquid biopsies search for around 10-20 mutations and up to around 100 in blood samples. Thus, they are able to detect ctDNA to levels on the range of one mutant molecule amongst 30,000 fragments of DNA. In contrast, the INVAR technique searched for hundreds and sometimes thousands of mutations in each blood sample, routinely achieving a sensitivity of one mutant molecule per 100,000, and under optimal conditions reached a level of ctDNA measured in parts per million.
The investigators applied INVAR to custom hybrid-capture sequencing data from 176 plasma samples from 105 patients with melanoma, lung, renal, glioma, and breast cancer across both early and advanced disease. By integrating signal across a median of more than 105 informative reads, ctDNA was routinely quantified to one mutant molecule per 100,000, and in some cases with high tumor mutation burden and/or plasma input material, to parts per million. This resulted in median area under the curve (AUC) values of 0.98 in advanced cancers and 0.80 in early-stage and challenging settings for ctDNA detection.
Senior author Dr. Nitzan Rosenfeld, senior group leader in cancer research at the University of Cambridge, said, “Personalized tests that can detect if cancer is still present, or find it early if it is returning, are now being tested in clinical trials. Whilst this may be several years away from clinical use, our research shows what is possible when we push such approaches to an extreme. It demonstrates that the levels of sensitivity we have come to accept in recent years in relation to testing for ctDNA can be dramatically improved. At present this is still experimental, but technology is advancing rapidly, and in the near future tests with such sensitivity could make a real difference to patients.”
The INVAR approach was described in the June 17, 2020 online edition of the journal Science Translational Medicine.
Related Links:
University of Cambridge









 (3) (1).png)