A Non-Invasive Urine Test for Prediction of Kidney Transplant Failure
By LabMedica International staff writers
Posted on 21 Apr 2020
A non-invasive test that predicts potential kidney transplant failure combines the CRISPR gene editing tool with a lateral-flow visual readout procedure to analyse urine samples from transplantation recipients.Posted on 21 Apr 2020
Investigators at the Max Delbrück Center for Molecular Medicine (Berlin, Germany) designed a novel assay system that screens for two common opportunistic viruses infecting kidney transplant patients, cytomegalovirus (CMV) and BK polyomavirus (BKV), and for CXCL9 mRNA, whose expression increases during acute cellular kidney transplant rejection.
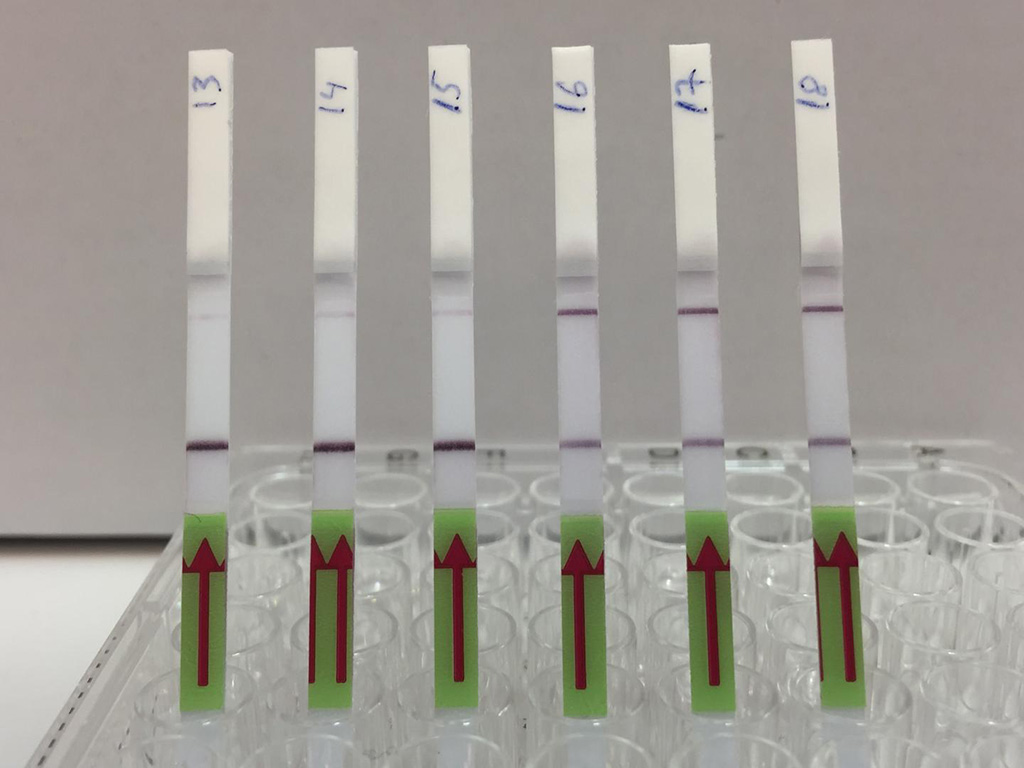
Image: The lateral flow strips show three patient samples that are negative for BK virus (13,14,15) and three patient samples that are positive (16,17,18). Presence of the upper band indicates a positive test result (Photo courtesy of Dr. Michael Kaminski, Max Delbrück Center for Molecular Medicine)
The first stage of the assay amplifies the DNA or RNA in the urine sample so that CRISPR can detect it. CRISPRs (clustered regularly interspaced short palindromic repeats) are segments of prokaryotic DNA containing short repetitions of base sequences. Each repetition is followed by short segments of "spacer DNA" from previous exposures to a bacterial virus or plasmid. Since 2013, the CRISPR/Cas9 system has been used in research for gene editing (adding, disrupting, or changing the sequence of specific genes) and gene regulation. By delivering the Cas9 enzyme and appropriate guide RNAs (sgRNAs) into a cell, the organism's genome can be cut at any desired location. The conventional CRISPR/Cas9 system from Streptococcus pyogenes is composed of two parts: the Cas9 enzyme, which cleaves the DNA molecule and specific RNA guides that shepherd the Cas9 protein to the target gene on a DNA strand.
Recent computational efforts to identify new CRISPR systems uncovered a novel type of RNA targeting enzyme, Cas13. The diverse Cas13 family contains at least four known subtypes, including Cas13a (formerly C2c2), Cas13b, Cas13c, and Cas13d. Cas13a was shown to bind and cleave RNA, protecting bacteria from RNA phages and serving as a powerful platform for RNA manipulation. It was suggested that Cas13a could function as part of a versatile, RNA-guided RNA-targeting CRISPR/Cas system holding great potential for precise, robust, and scalable RNA-guided RNA-targeting applications.
Following the DNA/RNA amplification step, the investigators applied the SHERLOCK protocol from SHERLOCK Biotechnologies (Cambridge, MA, USA). This is a method for single molecule detection of nucleic acid targets and stands for Specific High Sensitivity Enzymatic Reporter unLOCKing. It works by amplifying genetic sequences and programming a CRISPR molecule to detect the presence of a specific genetic signature in a sample, which can also be quantified. When it finds those signatures, the CRISPR enzyme is activated and releases a robust signal. This signal can be adapted to work on a simple paper strip test, in laboratory equipment, or to provide an electrochemical readout that can be read with a mobile phone.
Lateral flow analysis using a paper strip was used to convert the contents of the treated sample into an easily visualized result. One line appearing on the strip was a negative result, while two lines indicated a that virus is present. A similar process was used for CXCL9. MRNA was isolated and amplified, followed by CRISPR/Cas13 mediated target detection.
The investigators used the assay system to analyze more than 100 samples from kidney transplant patients. Results revealed that the test accurately detected BK polyomavirus DNA and cytomegalovirus DNA from patient-derived blood and urine samples, as well as CXCL9 messenger RNA at elevated levels in urine samples from patients experiencing acute kidney transplant rejection.
"Most people think of gene editing when they think of CRISPR, but this tool has great potential for other applications, especially cheaper and faster diagnostics," said first author Dr. Michael Kaminski, head of the kidney cell engineering and CRISPR diagnostics laboratory at the Max Delbrück Center for Molecular Medicine. "The challenge is getting down to concentrations that are clinically meaningful. It really makes a huge difference if you are aiming for a ton of synthetic target in your test tube, versus if you want to get to the single molecule level in a patient fluid."
The kidney transplant rejection assay was described in the April 13, 2020, online edition of the journal Nature Biomedical Engineering.
Related Links:
Max Delbrück Center for Molecular Medicine
SHERLOCK Biotechnologies