Plasma Virome of Brazilians with Unexplained Symptoms of Infection
By LabMedica International staff writers
Posted on 25 Mar 2020
Deep sequencing of nucleic acids in clinical samples now allows the identification of any known infectious agents, resulting in improved diagnostic capabilities. Blood from healthy people with high exposure to viral infections has also been analyzed using metagenomics. Posted on 25 Mar 2020
Analyzing plasma from patients with symptoms of acute viral infections such as fever of unknown origin may also be used as a surveillance tool for unexpected or novel (previously uncharacterized) viruses. Metagenomic analyses of plasma from patients with unexplained fever have also led to the characterization of previously unknown viral genomes.
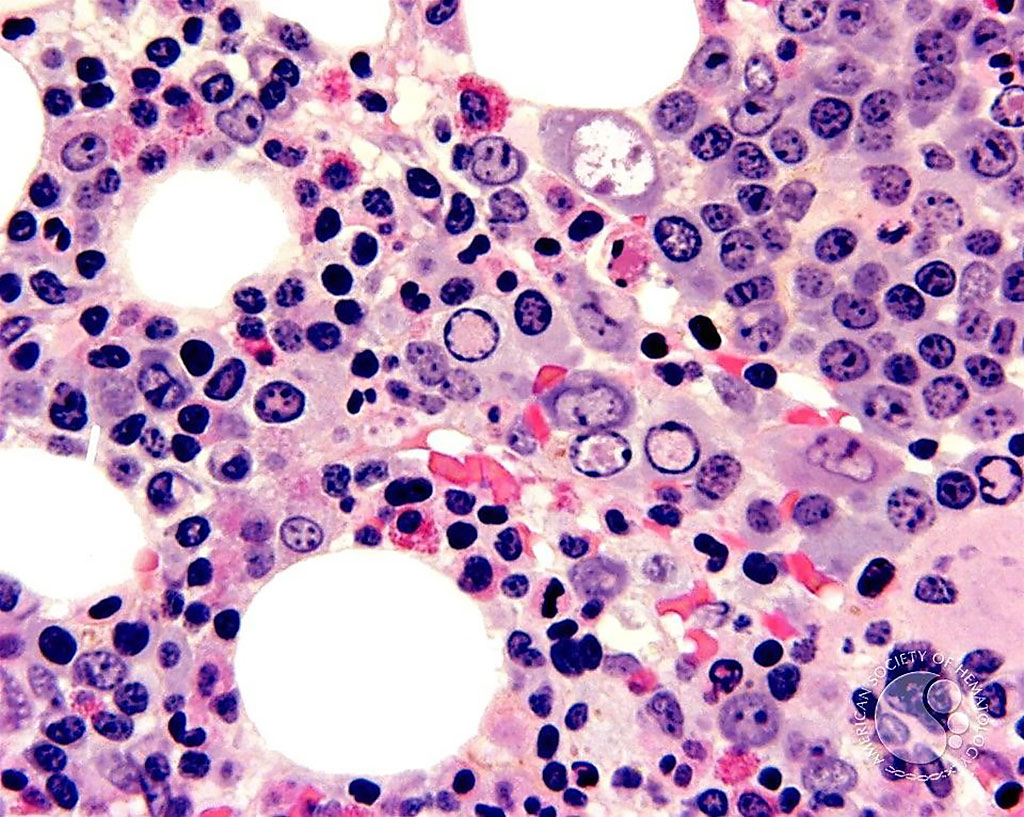
Image: Histological micrograph of a bone marrow biopsy from a patient with parvovirus. The parvovirus nuclear inclusions (clear areas) in the erythroblasts are more evident (Photo courtesy of John Lazarchick, MD).
A team of scientists collaborating with the Vitalant Research Institute (San Francisco, CA, USA) collected plasma from patients with dengue-like symptoms in 2013 to 2016 from the Brazilian states of Tocantins and Amapa. In this study, 781 samples tested negative for IgM against Dengue, Zika, and Chikungunya viruses and for flaviviruses, alphaviruses and enteroviruses RNA using RT-PCRs that were analyzed using viral metagenomics. Viral particles-associated nucleic acids were enriched, randomly amplified, and deep sequenced in 102 mini-pools generating over two billion reads. Sequence data was analyzed for the presence of known and novel eukaryotic viral reads. For the detection of viral RNAs a ZDC (Zika, Dengue, Chikungunya viruses) multiplex qPCR assay (Bio-Rad Laboratories, Inc.; Hercules, CA, USA) was used.
The investigators reported that Anelloviruses were detected in 80%, human pegivirus 1 in 19%, and parvovirus B19 in 17% of plasma pools. HIV and enteroviruses were detected in two pools each. Previously uncharacterized viral genomes were also identified, and their presence in single plasma samples confirmed by PCR. Chapparvovirus and ambidensovirus genomes, both in the Parvoviridae family, were partially characterized showing 33% and 34% identity in their NS1 sequences to their closest relative.
The team concluded that molecular surveillance using pre-existing plasma from febrile patients provides a readily scalable approach for the detection of novel, potentially emerging, viruses. Further testing the human tropism of the human plasma-associated parvovirus and densovirus reported will require detection of specific antibody responses, viral amplification in human cells, and/or the detection of viral RNA in cells of infected tissues. The study was published on March 5, 2020 in the journal PLOS ONE.
Related Links:
Vitalant Research Institute
Bio-Rad Laboratories