Real-Time PCR Developed for Eumycetoma Diagnosis
By LabMedica International staff writers
Posted on 06 Feb 2020
Mycetoma, a progressive and disfiguring disease, is one of the neglected tropical diseases, caused by both bacteria and fungi. Eumycetoma is the fungal type and mainly caused by species of the genus Madurella.Posted on 06 Feb 2020
There are four species of Madurella that are phenotypically similar and cause an invariable clinical picture, but differ markedly in their susceptibility to antifungal drugs, and epidemiological pattern. Therefore, specific identification is required for optimal management of Madurella infection and to reveal proper epidemiology of the species.
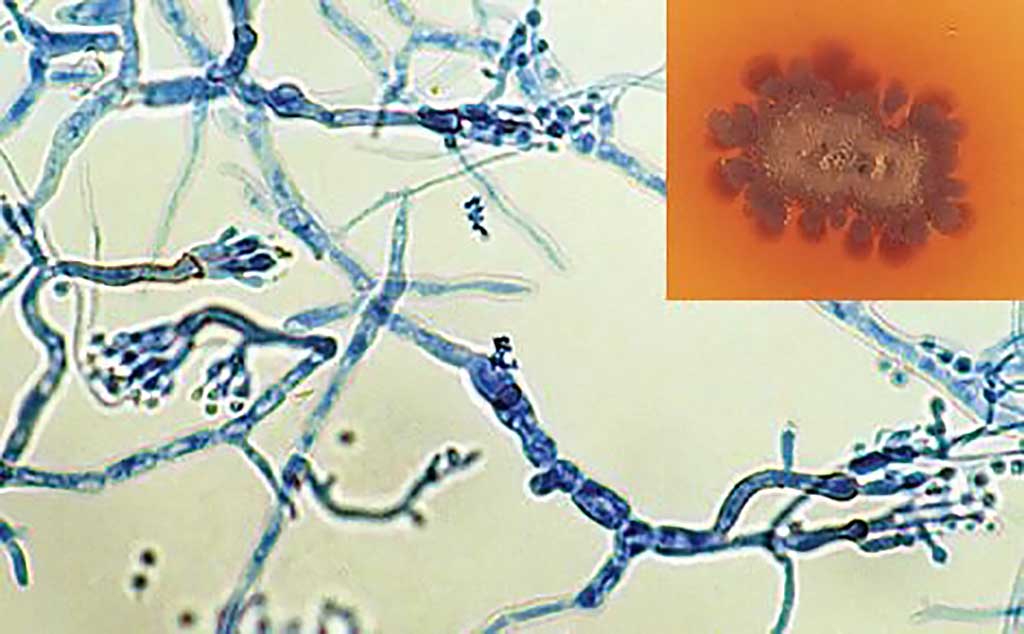
Image: Culture showing the typical brown diffusable pigment in the agar (inset) and phialides of Madurella mycetomatis (Photo courtesy of University of Adelaide).
Scientists from several countries cooperating with the Westerdijk Fungal Biodiversity Institute (Utrecht, The Netherlands) developed and standardized a novel multiplex real-time polymerase chain reaction (PCR) targeting the four Madurella species. In total 47 strains were included in this study, of which 26 belonged to the genus Madurella, nine to other members of the family Chaetomiaceae, nine to black-grain mycetoma species in the order Pleosporales, two to Aspergillus and one Rhizopus species. Thirteen clinical samples were obtained from patients seen at the Mycetoma Research Centre (Khartoum, Sudan)
Strains were grown on 2% Malt Yeast Extract agar (MEA) or Sabouraud Glucose Agar (SGA) plates and incubated for two weeks and mycelia were harvested and DNA was extracted. Purity of extracted DNA was assessed using NanoDrop ND-1000 spectrophotometer (Thermo Fisher Scientific, Wilmington, USA) and on 1% agarose gels. Polymerase chain reactions (PCR) were performed using an ABI 7500 fast PCR device (Applied Biosystems, Foster City, CA, USA).
The evaluation of the assay using reference strains of the target and non-target species resulted in 100% specificity, high analytical reproducibility, and a lowest detection limit of 3 pg target DNA. The accuracy of the real-time PCR was further assessed using biopsies from eumycetoma suspected patients. Unlike culture and DNA sequencing as gold standard diagnostic methods, the real-time PCR yielded accurate diagnosis with specific identification of the causative species in three hours compared to one or two weeks required for culture.
The authors concluded that they had developed a rapid and accurate diagnostic assay that is able to simultaneously identify M. fahalii, M. mycetomatis, M. pseudomycetomatis, and M. tropicana. The assay can be used in both culture-dependent and -independent manners. The novel method reduces turnaround time as well as labor intensity and high costs associated with current reference methods. The study was published on January 15, 2020 in the journal PLOS Neglected Tropical Diseases.
Related Links:
Westerdijk Fungal Biodiversity Institute
Mycetoma Research Centre
Thermo Fisher Scientific
Applied Biosystems