Alzheimer's Disease Gene Expression Varies by Brain Cell Type
By LabMedica International staff writers
Posted on 09 Dec 2019
Genetic variants in apolipoprotein E (APOE) and other genes have been implicated in late-onset Alzheimer's disease through genome-wide association studies and other analyses.Posted on 09 Dec 2019
There is currently little information available about how individual cell types contribute to Alzheimer’s disease and about the relative contributions that different cell subpopulations make to Alzheimer's disease risk and development, despite evidence pointing to a potential role for genes and pathways involved in endocytosis, microglial function, and neuronal connectivity.
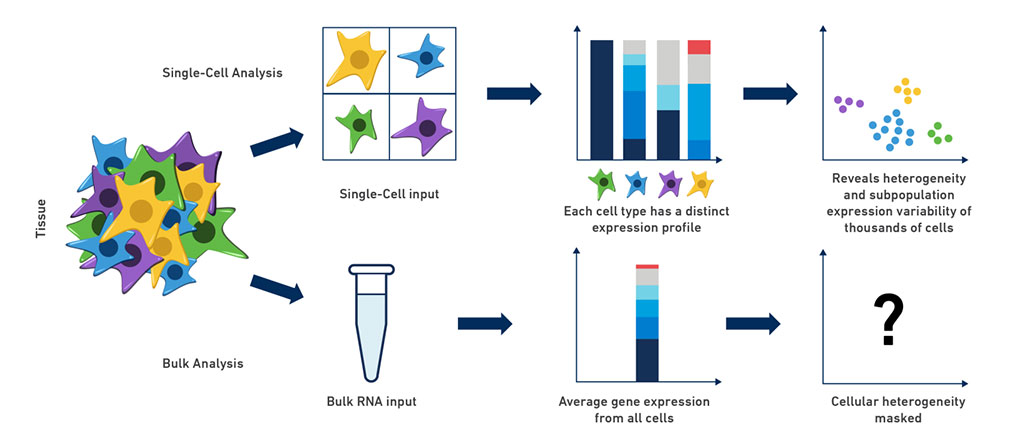
Image: Single-cell RNA-seq reveals cellular heterogeneity that is masked by bulk RNA-seq methods (Photo courtesy of 10x Genomics)
Scientists at Monash University (Clayton, Australia) applied single-nucleus RNA sequencing to entorhinal cortex samples from control and Alzheimer’s disease brains, yielding a total of 13,214 high-quality nuclei. The team used a DroNc-Seq protocol with the 10x Genomics platform (Pleasanton, CA, USA) to generate high-quality nuclear RNA sequences for 13,214 cells from entorhinal cortex samples from six individuals with Alzheimer's disease and six unaffected controls matched for age and sex.
The team identified cell clusters that roughly coincided with microglia, astrocyte, neuron, oligodendrocyte progenitor cell, oligodendrocyte, and endothelial cell types by combining the single-cell transcriptomes in combination with spatial information produced using an approach known as ‘uniform manifold approximation and projection.’ The investigators reported that the Alzheimer's disease-related gene APOE was found at enhanced levels in a certain subpopulation of microglia cells, for example, but was dialed down in nuclei from other brain cell subpopulations, including the oligodendrocyte progenitor and astrocyte cell populations.
The scientists reported that the Alzheimer’s disease risk gene APOE is specifically repressed in Alzheimer’s disease oligodendrocyte progenitor cells and astrocyte subpopulations and upregulated in an Alzheimer’s disease-specific microglial subpopulation. Integrating transcription factor regulatory modules with Alzheimer’s disease risk loci revealed drivers of cell-type-specific state transitions towards Alzheimer’s disease. For example, transcription factor EB, a master regulator of lysosomal function, regulates multiple disease genes in a specific Alzheimer’s disease astrocyte subpopulation.
The authors concluded that in light of the complex genetic contributions to late-onset Alzheimer's disease, it is important to understand late-onset Alzheimer's disease, genome wide association studies (GWAS) gene involvement in cell-type-specific transcription factor networks that drive the transitions of cells from health to Alzheimer's disease states. The study was published on November 25, 2019 in the journal Nature Neuroscience.
Related Links:
Monash University
10x Genomics