Triple-Negative Breast Cancer Classified from Whole-Genome Sequencing
By LabMedica International staff writers
Posted on 17 Oct 2019
A diagnosis of triple negative breast cancer means that the three most common types of receptors known to fuel most breast cancer growth–estrogen, progesterone and the HER-2/neu gene are not present in the cancer tumor.Posted on 17 Oct 2019
Triple-negative breast cancer (TNBC) can be more aggressive and difficult to treat. Also, the cancer is more likely to spread and recur. The stage of breast cancer and the grade of the tumor will influence the prognosis. The five-year survival rate for TNBC is around 77% versus 93% for other breast cancer types.
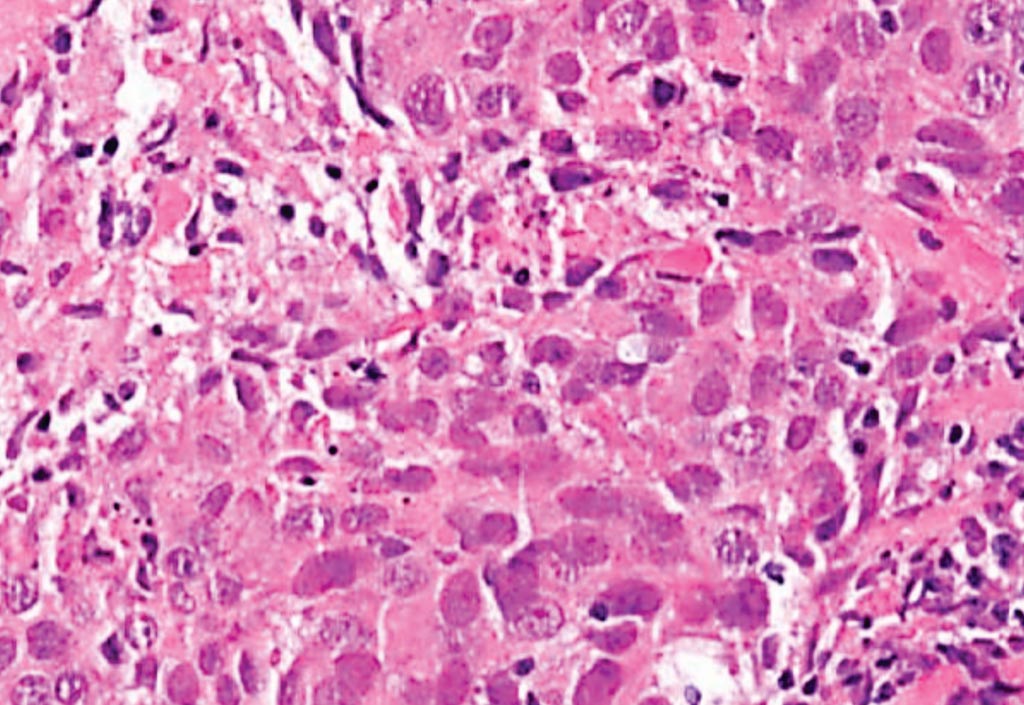
Image: A histopathology of high-grade invasive ductal carcinoma of the breast (not otherwise specified, grade 3). This is an example of a triple-negative breast cancer, basal-like carcinoma (Photo courtesy of Lee Moffitt Cancer Center).
An international team of scientists led by those at Lund University (Lund, Sweden) sequenced 254 TNBC tumors from patients from the Sweden Cancerome Analysis Network-Breast (SCAN-B) project, an ongoing population-based observational study of breast cancer patients in southern Sweden. By applying the previously developed machine-learning algorithm HRDetect to the genome sequences, the investigators divided the tumors into three groups: low, intermediate, and high, based on how deficient they were in homologous recombination-based gene repair. These groups had varying prognoses and responses to treatments.
After determining likely somatic drivers and pathogenic germline mutations for this cohort, the team applied the mutational signature-based algorithm HRDetect to examine the "BRCA-ness" or homologous recombination repair deficiency of these samples. Nearly 60% of the triple-negative tumors were classified as HRDetect-high; about 35% were HRDetect-low; and about 5% fell in an intermediate group. The scientists reported that the HRDetect-high group was enriched for patients under the age of 50 years, with high-grade tumors and a basal-like expression subtype. But as they noted, this group also included tumors with ER-staining, from middle-aged patients, and with non-basal-like gene expression profiles, suggesting that the HRDetect-high phenotype is enriched for the typical basal-like tumors found among young patients with triple-negative disease, but also encompasses others. This, they argued, suggests HRDetect represents a novel means of stratifying tumors.
Patients whose tumors fell into these different HRDetect groups also responded differently to treatment. Patients in the HRDetect-high group had the best outcomes of the three. These tumors were both more likely to respond to adjuvant chemotherapy as well as to PARP inhibitors. The HRDetect-intermediate group, meanwhile, had the worst outcomes, though the HRDetect-low group also had poor outcomes. But as the scientists noted, the molecular profiles of the HRDetect-intermediate and -low groups suggested potential actionable targets. The HRDetect-intermediate group, for instance, was enriched for tumors with Cyclin E1 (CCNE1) amplifications and for hypermutators of a rearrangement signature of long tandem duplications. The HRDetect-low group, meanwhile, was enriched for alterations to the PIK3CA/AKT1 pathway.
Johan Staaf, PhD, an associate professor of oncology and pathology, and first author of the study, said, “Using whole-genome sequencing, we can truly discriminate tumors that may or may not respond to current drugs among triple-negative breast cancer patients, a type of breast cancer that we still struggle to treat well.” The study was published on September 30, 2019, in the journal Nature Medicine.
Related Links:
Lund University