Genome-Wide Screening Identifies Pancreatic Cancer Biomarkers
By LabMedica International staff writers
Posted on 16 Apr 2019
Surgery currently offers the only chance for a cure in pancreatic ductal adenocarcinoma (PDAC), but it carries a significant morbidity and mortality risk and results in varying oncologic outcomes.Posted on 16 Apr 2019
PDAC accounts for 85% of pancreatic cancers and is one of the most malignant forms of the disease. One of the benefits of a biomarker signature is that single nucleotide polymorphism (SNP) genotyping can be determined at the time of diagnosis by blood test to identify patients at high risk for rapid tumor progression.

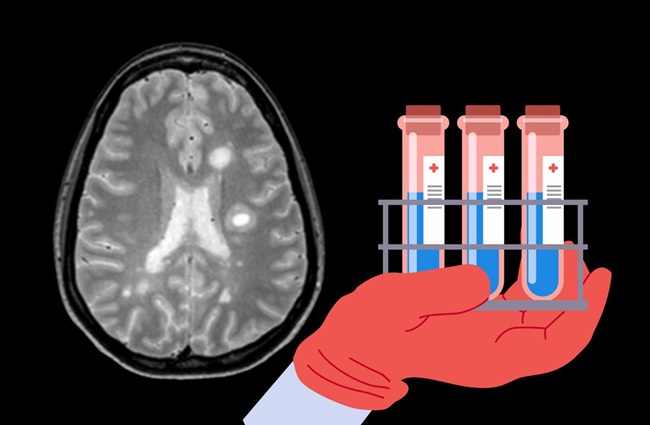
Image: The high-density Human Omni 2.5-8 SNP array (Photo courtesy of Illumina).
An international team of scientists collaborating with the University Hospital Zurich (Zurich, Switzerland) analyzed data from a prospective cohort of 195 PDAC cases in Europe who had undergone pancreatic resection looking for high-frequency SNPs associated with allelic differences in tumor-associated survival. They looked for SNPs that either alter protein structure or reside in known regulatory noncoding genomic regions. They then used data from 136 patients from the pancreatic cancer cohort in the Cancer Genome Atlas database to validate the findings.
To obtain an initial list of SNPs, the scientists genotyped over two million SNPs from the European cohort using Illumina's high-density Human Omni 2.5-8 SNP array. They pared down the list of variants using allele frequency information, and used the Ensembl Variant Effect Predictor software to find overlaps between SNP loci and known regulatory regions from publicly available genome-wide assays. They also looked for common coding SNPs that are known to have deleterious effects on protein function. The final list contained more than 38,000 SNPs.
The team reported two SNPs in noncoding, functional regions of genes that regulate cancer progression, invasion, and metastasis were identified (CHI3L2 SNP rs684559 and CD44 SNP rs353630). These were associated with survival after PDAC resection; patients who carry the risk alleles at one of both SNP loci had a 2.63-fold increased risk for tumor-associated death compared with those with protective genotypes (hazard ratio for survival = 0.38).
The authors concluded that they the identified polymorphisms may serve as a noninvasive biomarker signature of prospective survival after pancreatic resection that is readily available at the time of PDAC diagnosis. The signature could be used to identify high-risk PDAC patients with a low probability of survival that might be candidates for clinical trials of new therapeutic strategies including neoadjuvant chemotherapy protocols. They also note that these SNPs could aid in the development of individualized genomic strategies for PDAC therapies. The study was published on April 3, 2019, in the journal JAMA Surgery.
Related Links:
University Hospital Zurich