Targeted Sequencing Approach for Sarcoma Gene Fusions Validated
By LabMedica International staff writers
Posted on 12 Sep 2018
Sarcomas are rare cancers that grow in a patient's connective tissue, such as bones, muscles, tendons, nerves, and fat. Symptoms may not be apparent, since the only signs of sarcomas are lumps on a person's skin. Posted on 12 Sep 2018
It has been demonstrated that recurrent gene translocations are prevalent in these cancers. Although scientists currently use a variety of techniques to detect translocations, they often use fluorescence in situ hybridization (FISH) in routine diagnostic settings. Despite its proven uses, the tool's drawbacks include false-negative results when there are a high percentage of non-tumor cells, atypical FISH signals, and the dependency of probe availability.
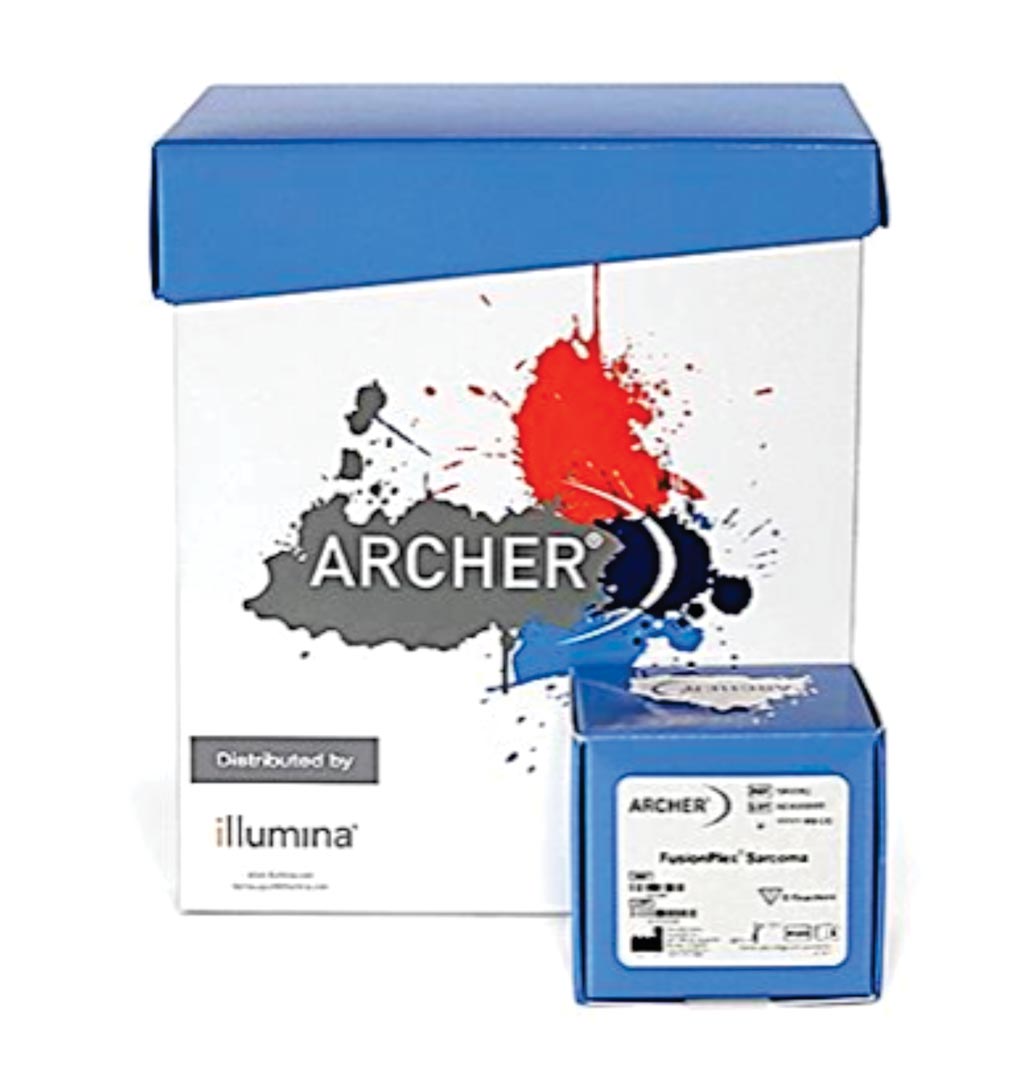
Image: The Archer FusionPlex Sarcoma Kit: A targeted sequencing assay to simultaneously detect and identify fusions of 26 genes associated with soft tissue cancers (Photo courtesy of Illumina).
Pathologists at the Leiden University Medical Center (Leiden, the Netherlands) collected 81 soft tissue or bone marrow samples, eight freshly frozen and 73 formalin-fixed, paraffin-embedded (FFPE), that either had a fusion-positive or unknown translocation status. To detect gene translocations, they isolated DNA and RNA from the tissue samples, and then prepared sequencing libraries of target-enriched complementary DNA (cDNA) using the FusionPlex Sarcoma kit (ArcherDX, Boulder, CO, USA).
The scientists used commercial FISH probes to detect translocations involving EWSR1, FUS, ALK, SS18, USP6, COL1A1 and PDGFB. In addition, the team used TaqMan probes to detect the most common fusion transcripts in Ewing and synovial sarcomas. The team performed immunohistochemistry for CAMTA1, TFE3, STAT6, ALK, ETV4, and CCNB3 mutations and then directly compared the AMP-based targeted outcomes to results from the secondary molecular techniques. A mixture of probes, primers, and cDNA were admixed with a qPCR Core kit.
The team reported that next-generation sequencing (NGS) yielded an average of 2.4 million read numbers per sample, with an average of 64% RNA reads. They found that 48 cases showed a fusion with AMP-based targeted NGS. In the remaining 33 cases, they did not find any gene fusion mutations. In addition, NGS results were in concord with FISH or RT-PCR in 29 of the cases. They included eight cases of Ewing sarcomas, with seven demonstrating an EWSRI-FLI1 fusion. In situations with discrepant results, the team pursued further analysis with additional FISH probes and RT-PCR primers. In seven samples, the team could not produce a specific immunohistochemical evaluation. However, they noted that these cases had histologic and immunohistochemical features that were characteristic for the tumors.
The team could not detect any specific fusion with ESWRI, despite demonstrating convincing ESWRI breakage with FISH probes. In contrast, the team found that both RT-PCR and FISH were easy to operate, inexpensive, and had high clinical accuracy. When using RT-PCR to make DNA, however, the team needed to ensure they had selected the correct translocation areas on the genome, as they might have to repeat the process if they identified the wrong fusion type.
Anne-Marie Cleton-Jansen, PhD, a study author and an associate pathology professor, said, “Regarding sarcomas and gene fusions, if a patient has a particular gene fusion, it's easier to perform a diagnosis. However, it must be done in the background of histology and patient specifics. One of the great things about using the FusionPlex Sarcoma kit is that you only need to know one of the translocation partners. You can then identify the second partner by next-generation sequencing.” The study was published on August 19, 2018, in The Journal of Molecular Diagnostics.
Related Links:
Leiden University Medical Center
ArcherDX