Current-Tunneling Measurements Analyze Single DNA Molecules
By LabMedica International staff writers
Posted on 10 Jul 2018
A team of Japanese genomics researchers has devised a method for cancer diagnosis, which is based on the analysis of single molecules of DNA without the need for chemical modification or amplification.Posted on 10 Jul 2018
Cancer can be diagnosed by identifying DNA and microRNA base sequences that have the same base length yet differ in a few base sequences, if the abundance ratios of these slightly deviant base sequences can be determined. However, such quantitative analyses cannot be performed using the current DNA sequencers.
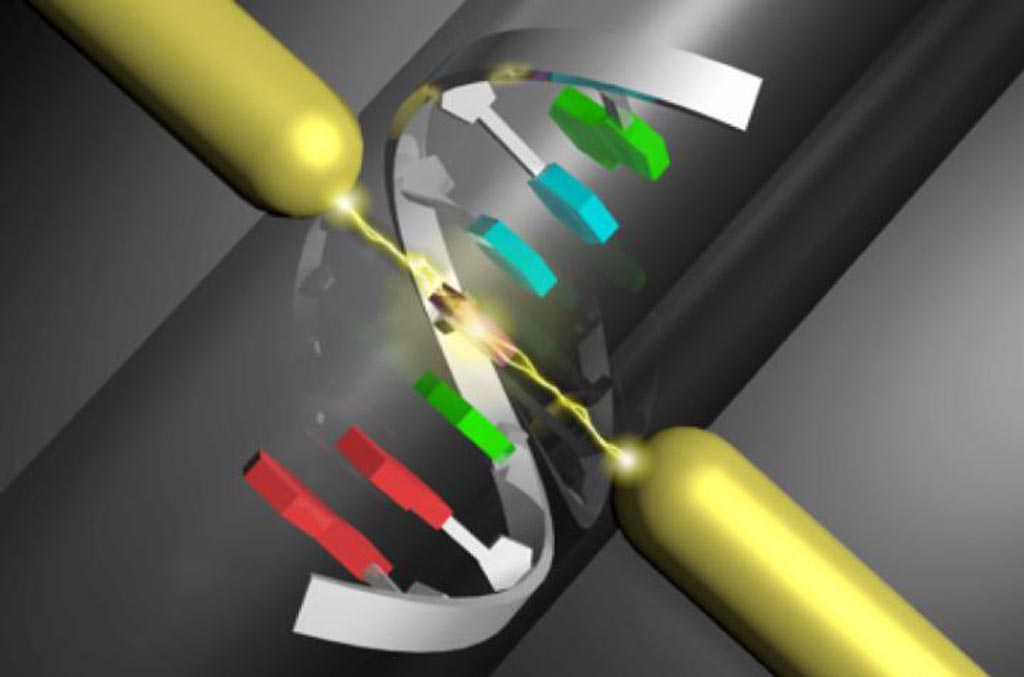
Image: An artist\'s depiction of the operating principle of single-molecule sequencing (Photo courtesy of Osaka University).
In this regard, investigators at Osaka University (Japan) used current-tunneling measurements to determine the entire base sequences of four types of DNA corresponding to the let-7 microRNA, which is a 22-base cancer marker.
The tunneling currents flowing through single molecules were measured by gold electrodes – maintained at a distance of 0.75 nanometers from each other, equivalent to the size of a DNA base molecule – using a mechanically controllable break-junction. Single-molecule signals were obtained in forms of current spikes, whose height represented the electron transport through the molecule. Since this method measured single molecules, it did not require chemical modification of DNA or amplification by PCR.
As the method measured individual DNA molecules, two or more base sequences could be determined by measuring a solution of DNA molecules with two or more types of base sequences. Furthermore, since this method could count the number of DNA molecules that contained a specific base sequence, quantitative analysis could detect the base sequences and determine their frequency.
"Because the single-molecule sequencing method detects differences in the electronic states of molecules in terms of single-molecule conductances, it may also be applied to the analysis of microRNA and RNA molecules that include four base molecules and peptides that include 20 kinds of amino acids," said senior author Dr. Masateru Taniguchi, a professor in the institute of scientific and industrial research at Osaka University. "Also, as the method can detect chemically modified base molecules and amino acids, it represents a substantive step toward realizing personalized genomic diagnosis of cancer and other diseases."
The current-tunneling method was described in a paper published in the June 4, 2018, online edition of the journal Scientific Reports.
Related Links:
Osaka University













