RT-PCR Assay Developed to Detect Bloodstream Infections
By LabMedica International staff writers
Posted on 11 Jun 2018
Rapid detection and identification of a causative pathogen is essential in the treatment of critically ill patients with blood stream infection (BSI), since timely initiation of adequate antibiotic treatment is associated with decreased morbidity, mortality, and possibly reduced healthcare costs.Posted on 11 Jun 2018
Conventional culture of inoculated blood samples, termed blood culture (BC), is currently considered the “gold standard” for diagnosing BSI. However, its diagnostic accuracy may be hampered by concomitant antibiotic treatment, low levels of circulating bacteria, and poor sensitivity for slow growing, intracellular, and fastidious microorganisms.
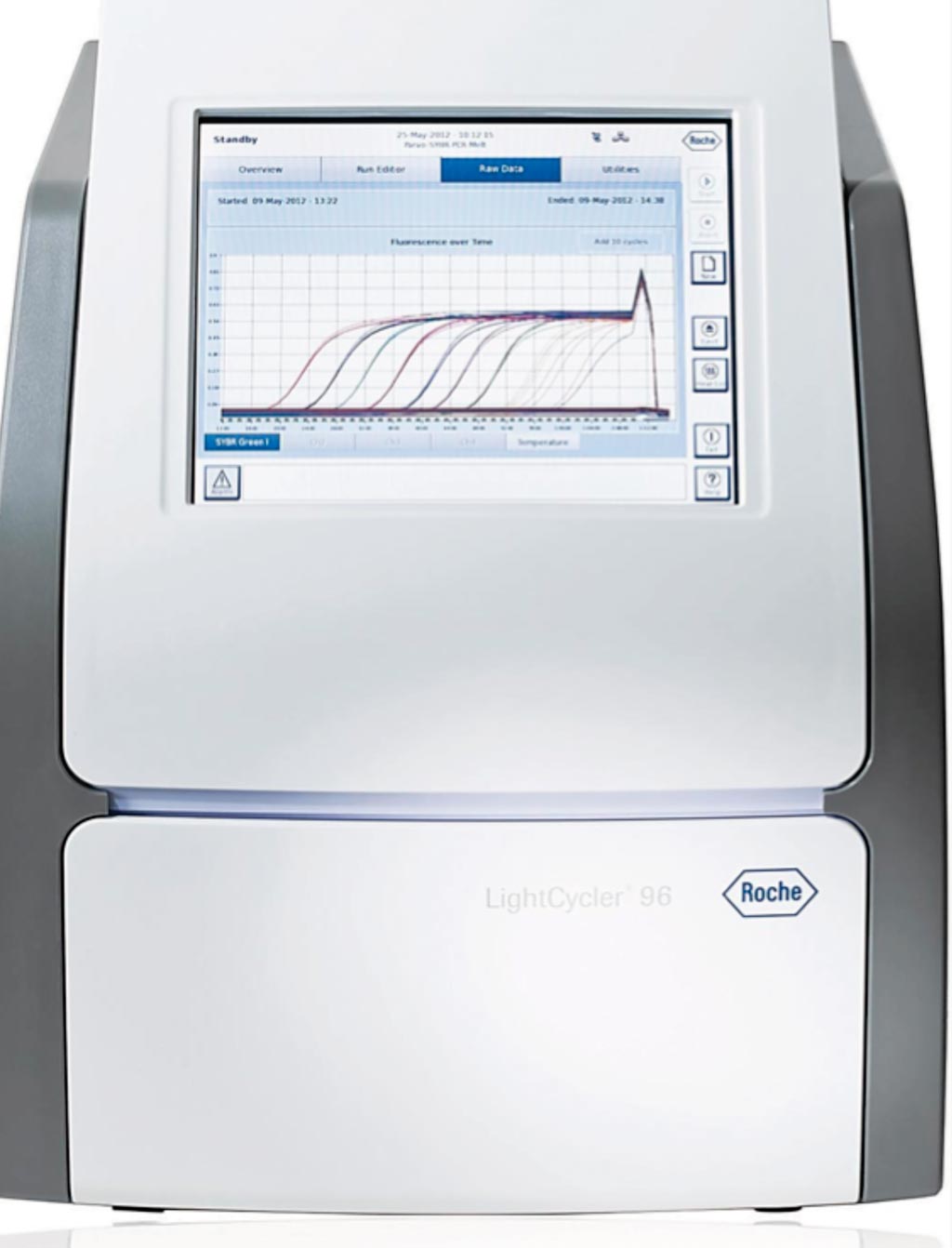
Image: The LightCycler 96 real-time PCR system (Photo courtesy of Roche Diagnostics).
Scientists at the University of Utrecht Medical Center (Utrecht, the Netherlands) and their colleagues collected 5 mL of blood from critically ill patients across multiple years for both blood culture and polymerase chain reaction (PCR) testing. They used 347 blood-culture positive samples, representing up to 50 instances for each pathogen covered by the assay, as well as 200 blood-culture negative samples in order to compare PCR results. After sample collection, the team added a buffer solution and performed centrifugation on the samples, isolating 7 mL to 10 mL of pathogen DNA per sample for PCR testing.
The team designed three novel multiplex assays in order to detect specific pathogens at the species level, as well as an additional broad PCR assay, called molecular Gram stain, to discriminate clinically relevant Gram-negative specimens from Gram-positive specimens. Bacterial pathogens included Escherichia coli, Enterococcus faecium, E. faecalis, Acinetobacter baumannii, and Staphylococcus aureus. In addition, the researchers included probes for Candida species, Aspergillus, and the resistance markers mecA and CTX- M1,9.
The blood stream infection (BSI-PCR) assays were run on a LightCycler system. Bacterial species-specific PCR sensitivities ranged from 65% to 100%. Sensitivity was 26% for the Gram-positive PCR, 32% for the Gram-negative PCR, and ranged 0% to 7% for yeast PCRs. Yeast detection was improved to 40% in a smaller set-up. There was no overall association between BSI-PCR sensitivity and time-to-positivity of BC (which was highly variable), yet Ct-values were lower for true-positive versus false-positive PCR results. False-positive results were observed in 84 (4%) of the 2,200 species-specific PCRs in 200 culture-negative samples, and ranged from 0% to 6% for generic PCRs.
The authors concluded that that there was no overall link between BSI-PCR sensitivity and time to positivity of blood culture. Overall, they believe that sensitivity of the BSI-PCR is promising for individual bacterial pathogens, but still inadequate for yeasts and generic PCRs. The study was originally published online on April 26, 2018, in the European Journal of Clinical Microbiology and Infectious Diseases.
Related Links:
University of Utrecht Medical Center