New Gene Mutation Associated with Fanconi Anemia
By LabMedica International staff writers
Posted on 25 Jul 2017
Fanconi anemia is a rare genetic disease characterized by bone marrow failure heralded by low platelet counts and unusually large red blood cells. Mutations in over 20 genes have been identified as causative for Fanconi anemia, which encode proteins commonly involved in DNA repair mechanisms.Posted on 25 Jul 2017
Safeguarding of the genome is essential for the suppression of oncogenesis and for stem cell maintenance in many organisms, and is warranted by DNA repair mechanisms. If they fail on a genetic basis, DNA repair disorders occur and Fanconi anemia (FA) is such a rare human genomic instability disease.
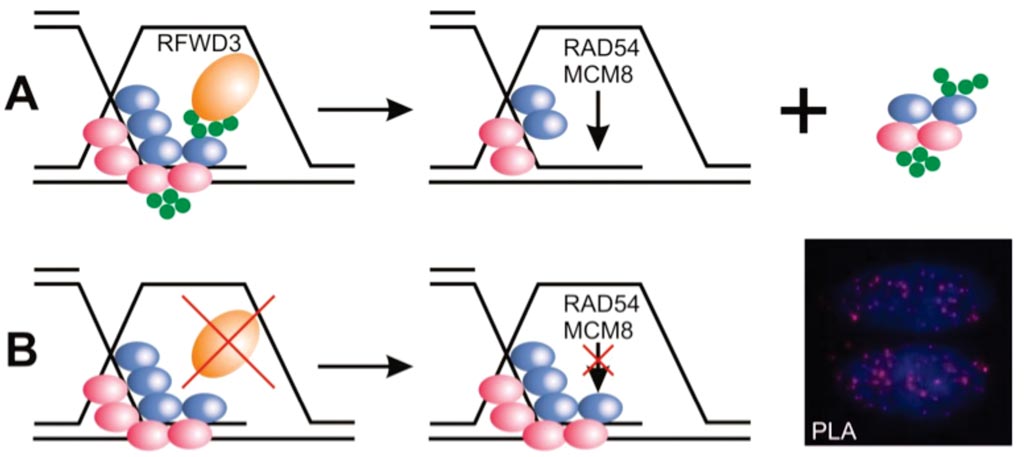
Image: The enzyme E3 ubiquitin ligase (RFWD3) helps target other proteins on single-stranded DNA for degradation. B: Cells lacking RFWD3 show DNA repair defects (Photo courtesy of Julius-Maximilians-Universität).
Scientists at the Julius-Maximilians-Universität (Würzburg, Germany) reports classical Fanconi anemia symptoms in a 12-year-old individual without mutations in any of the known Fanconi anemia genes. The patient presented with congenital abnormalities characteristic of FA and had a typical FA phenotype and compound heterozygous mutations in the E3 ubiquitin ligase (RFWD3).
The team used various techniques in the study including DNA sample preparation, polymerase chain reaction (PCR), Sanger sequencing, and whole exome sequencing, on a HighSeq2000 instrument. Other methods used were generation of stable lentiviral producer lines and transduction of adherent cells, DT40 gene targeting, cell cycle, cell survival, and chromosomal studies, where cell numbers were determined using a Nucleo Counter NC-250. Immunofluorescence studies were analyzed on a BIOREVO BZ-9000 microscope.
Sequencing of this individual's genome detected mutations in both alleles of the gene RFWD3, which encodes an enzyme that helps target other proteins on single-stranded DNA for degradation. This process is impaired in patient's cells, which rendered them more sensitive to chromosome breakage and DNA damage, compared to cells from healthy individuals. Other cells either lacking RFWD3 or genetically engineered with the patient's missense mutation showed similar DNA repair defects, which were rescued by expression of wild-type RFWD3. Together, these findings support the identification of RFWD3 as a Fanconi anemia gene. The study was published on July 10, 2017, in the Journal of Clinical Investigation.
Related Links:
Julius-Maximilians-Universität