Noninvasive Prenatal Testing of Single Gene Diseases Developed
By LabMedica International staff writers
Posted on 01 Feb 2017
An approach that allows for the noninvasive assessment of both maternally and paternally inherited mutations involves the analysis of single nucleotide polymorphisms (SNPs) in maternal plasma DNA with reference to parental haplotype information.Posted on 01 Feb 2017
In the past, parental haplotypes were resolved by complex investigational methods or inferential approaches, such as through the analysis of DNA from other affected family members. Recently, microfluidics-based linked-read sequencing technology has become available and allows the direct haplotype phasing of the whole genome rapidly.
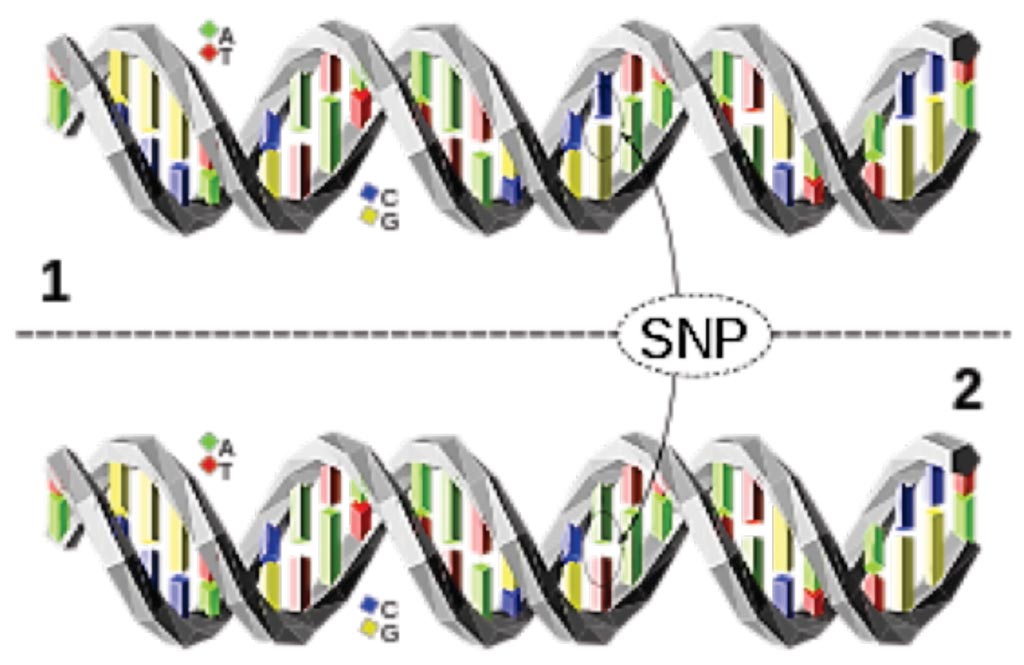
Image: A Single Nucleotide Polymorphism (SNP) is a change of a nucleotide at a single base-pair location on DNA. The upper DNA molecule differs from the lower DNA molecule at a single base-pair location (a C/A polymorphism) (Photo courtesy of David Eccles).
Scientists at the Chinese University of Hong Kong developed a test that can detect almost any single-gene disorder in the first six to 10 weeks of pregnancy. Guided by DNA from parent blood samples, the test looks for increases in the level of mutations associated with a particular condition in the mother’s blood once she is pregnant. In theory, any hospital pathology laboratory could run the test, producing results in one to two weeks.
The team first resolved the haplotypes of parental genomes with the use of linked-read sequencing technology. Then, they identified single nuclear polymorphisms (SNPs) within and flanking the genes of interest in maternal plasma DNA by targeted sequencing. Finally, they applied relative haplotype dosage analysis to deduce the mutation inheritance status of the fetus. They reported that haplotype phasing and relative haplotype dosage analysis of 12 out of 13 families were successfully achieved. The mutational status of these 12 fetuses was correctly classified.
The authors concluded that high-throughput linked-read sequencing followed by maternal plasma-based relative haplotype dosage analysis represents a streamlined approach for noninvasive prenatal testing of inherited single gene diseases. The approach bypasses the need for mutation-specific assays and is not dependent on the availability of DNA from other affected family members. Thus, the approach is universally applicable to pregnancies at risk for the inheritance of a single gene disease. The study was published in the January 2017 issue of the journal Clinical Chemistry.