Brain Infections Diagnosed Using Next-Generation Sequencing
By LabMedica International staff writers
Posted on 06 Jul 2016
The feasibility of next-generation sequencing (NGS) microbiome approaches in the diagnosis of infectious disorders in brain or spinal cord biopsies in patients with suspected central nervous system (CNS) infections has been investigated.Posted on 06 Jul 2016
This NGS technology can provide a view of the transcriptome of the host tissue as well as capture microbial genomes such as bacteria, fungi, and viruses that reside in the tissue niche. Deep sequencing of total DNA or ribonucleic acid (RNA) provides an unbiased approach that can detect even rare components of the microbiome.
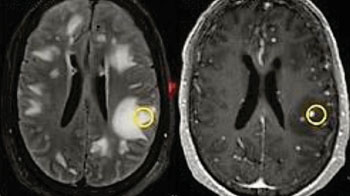
Image: An MRI scan of a brain lesion (white) in a patient with tuberculosis from the study. The yellow circle indicates the biopsy location (Photo courtesy of Johns Hopkins University).
A team of scientists at Johns Hopkins University (Baltimore, MD, USA) performed a prospective pilot study, and applied NGS in combination with a new computational analysis pipeline to detect the presence of pathogenic microbes in brain or spinal cord biopsies from 10 patients with neurologic problems indicating possible infection, but for whom conventional clinical and microbiology studies yielded negative or inconclusive results. Fresh frozen tissues from eight cases were sequenced immediately after biopsy and two other samples were from paraffin-processed tissues.
The team found that direct DNA and RNA sequencing of brain tissue biopsies generated 8.3 million to 29.1 million sequence reads per sample, which successfully identified with high confidence the infectious agent in three patients for whom validation techniques confirmed the pathogens identified by NGS. Although NGS was unable to identify with precision infectious agents in the remaining cases, it contributed to the understanding of neuropathological processes in five others, demonstrating the power of large-scale unbiased sequencing as a novel diagnostic tool. Clinical outcomes were consistent with the findings yielded by NGS on the presence or absence of an infectious pathogenic process in eight of 10 cases, and were noncontributory in the remaining two.
Carlos Pardo-Villamizar, MD, an associate professor of neurology, a senior author of the study, said, “By incorporating modern genetic sequencing techniques into pathology diagnostics, we were able to investigate the potential presence of infection in 10 subjects and found appropriate explanations of clinical problems in eight out of 10 patient cases examined in this study. We hope to develop this technique further as a way to bring the diagnosis rate of inflammatory brain disorders and infections closer to 100% so we can treat patients more effectively.” The study was published on June 13, 2016, in the journal Neurology: Neuroimmunology & Neuroinflammation.
Related Links:
Johns Hopkins University