New Method to Reveal Bacterial Reaction to Antibiotics in Five Minutes Could Help Create Rapid Molecular Test
Posted on 24 May 2023
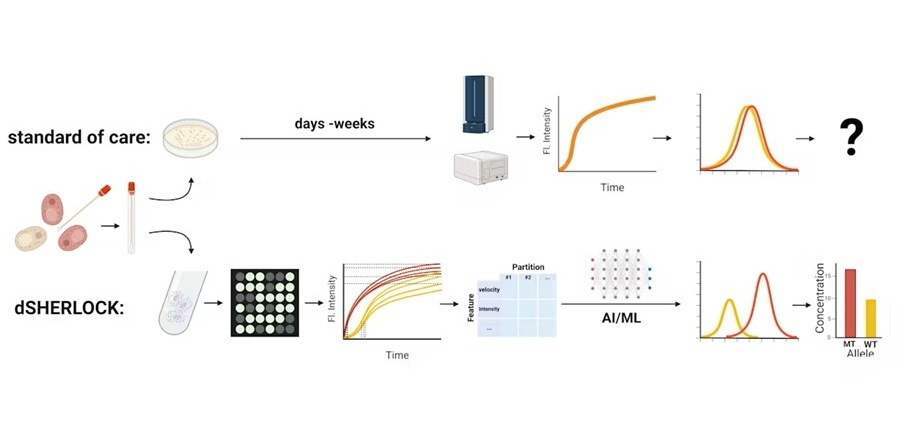
Severely sick patients suffering from bacterial infections often require immediate treatment to prevent serious health complications, making it vital for physicians to quickly identify the appropriate antibiotic. However, existing approaches to determining antibiotic resistance can involve extensive periods, sometimes hours or even days. This has led to the frequent prescription of broad-spectrum antibiotics, heightening the risk of antibiotic resistance. Now, a simple method has been developed that can detect bacterial response to antibiotics within just five minutes.
Researchers at Karolinska Institutet (Solna, Sweden) set out to reduce the unwarranted use of antibiotics by devising a rapid method to assess how bacteria react to different environmental conditions, including antibiotic administration. They developed the 5PSeq method, which relies on sequencing the messenger RNA (mRNA) that is broken down by the bacteria as they synthesize proteins. The researchers employed the 5PSeq method to examine mRNA breakdown intermediates in isolated species and complex microbiomes. They tested the method on 96 bacterial species from diverse phyla in complex clinical samples, such as fecal, gut, and vaginal samples, as well as compost samples. In a matter of minutes, the researchers were able to determine whether the bacteria were reacting to the antibiotic treatment; the effect was most noticeable after about half an hour.

By utilizing metadegradome sequencing - parallel analysis of RNA ends - the team characterized 5′P mRNA decay intermediates in all 96 species, including Bacillus subtilis, Escherichia coli, Synechocystis spp., and Prevotella copri. They discovered co-translational mRNA degradation to be common among bacteria and generated a degradome atlas for the 96 species, facilitating the further study of RNA degradation mechanisms in bacteria. In addition to measuring antibiotic resistance, the method can be employed to help scientists understand how bacteria manage diverse environmental pressures, and how they interact both with each other and with their hosts. The researchers plan to continue investigating complex intestinal samples to gain deeper insights into the interactions of bacterial communities in the gut and their effects on human health. The aim is to refine the method and develop a rapid molecular test for clinical application.
“We demonstrate that metadegradome sequencing provides fast, species-specific posttranscriptional characterization of responses to drug or environmental perturbations,” the researchers wrote. “Our work paves the way for the application of metadegradome sequencing to investigation of posttranscriptional regulation in unculturable species and complex microbial communities.”
Related Links:
Karolinska Institutet