Detection of Pathogenic Microorganisms in Semen by Sequencing Sperm RNA
By LabMedica International staff writers
Posted on 11 Feb 2020
Advanced sequencing of sperm RNA was used to detect genetic material from microorganisms that were present as contaminants in the associated semen.Posted on 11 Feb 2020
The current approach for analysis of the male reproductive tract microbiome relies on identification of micoroorganisms grown from cultures. This approach is of limited value, since the majority of pathogens cannot be cultured.
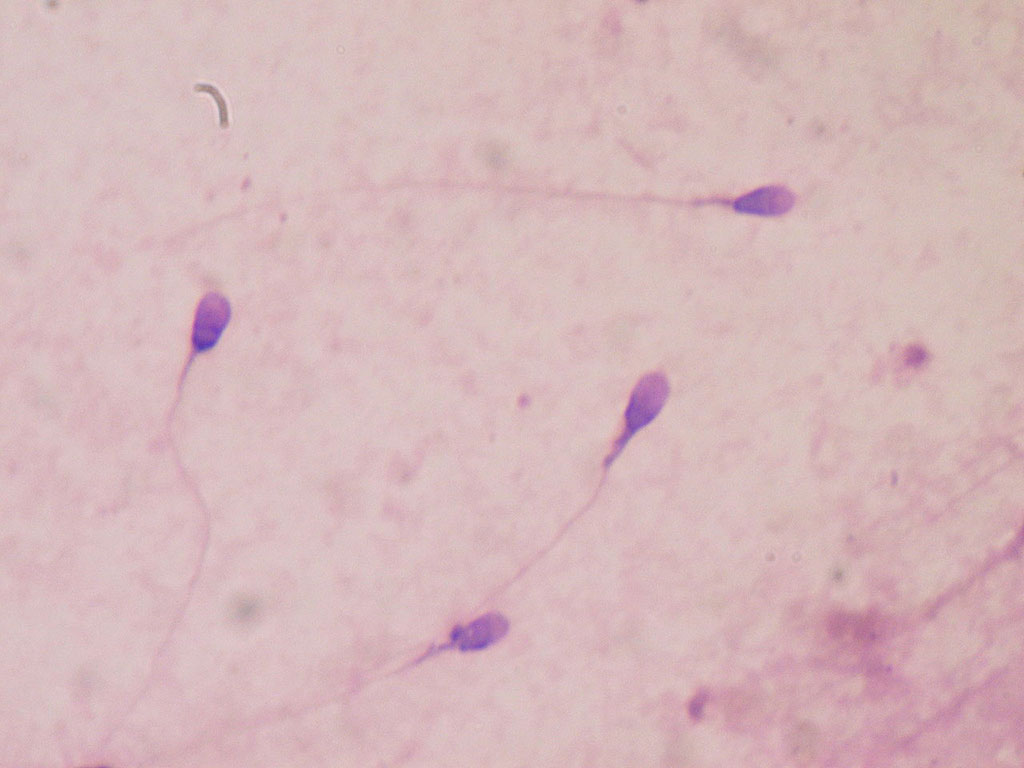
Image: Human sperm stained for semen quality testing in the clinical laboratory (Photo courtesy of Wikimedia Commons)
Investigators at Wayne State University (Detroit, MI, USA) and colleagues at the CReATe Fertility Center (Toronto, Canada) and the University of Massachusetts Amherst (USA) conducted a study designed to assess the capacity of human sperm RNA-seq (RNA sequencing) data to gauge the diversity of the microbiome present within the semen.
For this study, the investigators collected 85 semen samples, isolated the sperm RNA, and analyzed the material using RNA sequencing technology. Microbial composition was determined by aligning sequencing reads not mapped to the human genome to the [U.S.] National Center for Biotechnology Information (NCBI) RefSeq bacterial, viral, and archaeal genomes.
Microbial composition within each sample was characterized as a function of microbial associated RNAs. Results revealed that bacteria known to be associated with the male reproductive tract were present at similar levels in all samples representing 11 genera from four phyla with one exception, an outlier. The outlier sample exhibited a dramatic increase in Streptococcus, represented by two operational taxonomic units, S. agalactiae and S. dysgalactiae. These bacteria cause neonatal infection during pregnancy and post-delivery and are linked to significant mortality rates in premature births.
"We show that non-targeted sequencing of human sperm RNA has the potential to provide a profile of micro-organisms (bacteria, viruses, archaea)," said Dr. Stephen Krawetz, professor of fetal therapy and diagnosis at Wayne State University. "This information was recovered from the data typically cast aside as part of routine nucleic acid sequencing. The enhanced sensitivity and specificity of the sequencing technology as compared to current approaches may prove useful as a diagnostic tool for microbial status as part of the routine assessment as we move toward personalized care. Given the recent increase and severity of Streptococcus (agalactiae) infection, as well as others in adults, neonates, and newborns, non-targeted human sperm RNA sequencing data may, in addition to providing fertility status, prove useful as a diagnostic for microbial status."
The RNA sequencing study was published in the January 4, 2020, online edition of the Journal of Assisted Reproduction and Genetics.
Related Links:
Wayne State University
CReATe Fertility Center
University of Massachusetts Amherst