Fungus-Associated Bacteriome Associated with C. difficile Infection
By LabMedica International staff writers
Posted on 12 Sep 2019
Clostridioides difficile infection (CDI), the most common etiology of nosocomial infectious diarrhea, is caused by an intestinal dysbiosis that is virtually always attributable to antibiotics. This causes one of the most common hospital-acquired infections in the USA.Posted on 12 Sep 2019
There is a growing body of data showing that distinct bacterial and fungal community structures distinguish the dysbiotic state of CDI from antibiotic-associated diarrhea due to other causes. The data suggests that transkingdom interactions between fungal and bacterial species play an important role in nosocomial infections.
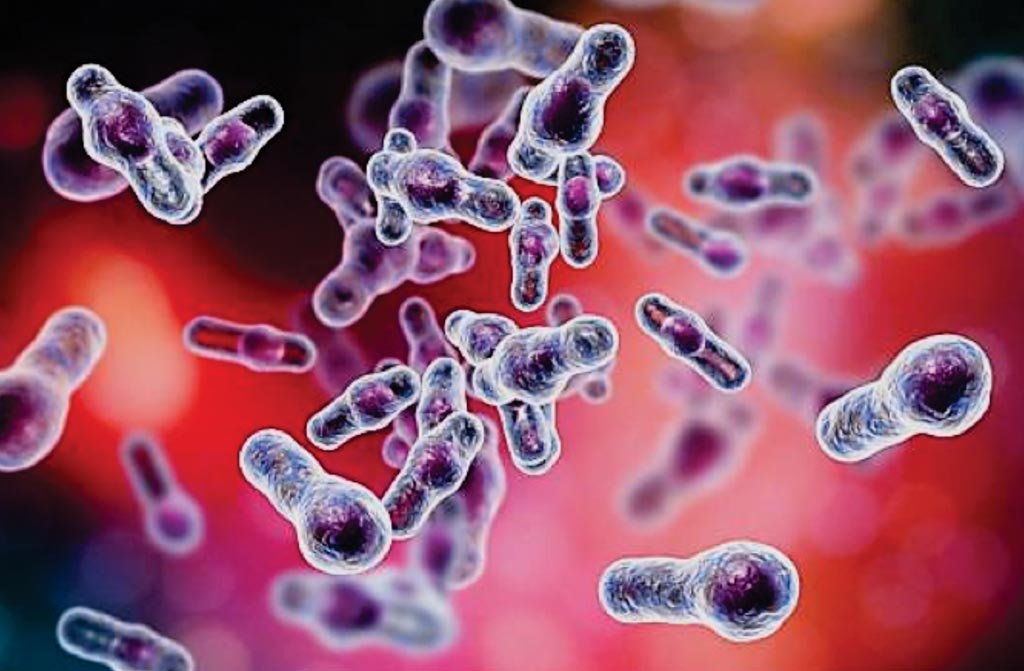
Image: Drumstick-shaped Clostridioides difficile bacilli as they appear in scanning electron microscopy (Photo courtesy of Kateryna Kon, PhD).
A team of scientists collaborating with the University of Arizona (Tucson, AZ, USA) collected diarrheal stools from 49 inpatients (18 of whom tested positive for CDI) under stringent inclusion criteria. C. difficile testing was performed using a commercially available nucleic acid amplification test designed to detect a highly conserved sequence within the tcdA gene. C. difficile positive and negative stool samples were preserved.
Fecal DNA extracts were subject to 16S rRNA gene and ITS2 Illumina tag polymerase chain reaction (PCR) (Illumina, San Diego, CA, USA; www.illumina.com), pooled in equimolar ratios, gel purified, and sequenced on the Illumina MiSeq (16S rRNA libraries) and NextSeq (ITS libraries) platforms. Bacterial (16S rRNA gene) and fungal (ITS) sequences were quality filtered, clustered into operational taxonomic units (OTUs), and normalized using both the USEARCH and QIIME pipelines. The team utilized a tiered sequencing approach to identify enriched bacterial and fungal taxa, using 16S and internal transcribed spacer (ITS) rRNA gene amplicon sequencing, with matched metagenomics and metatranscriptomics performed on a subset of the population.
The scientists reported that distinct bacterial and fungal compositions distinguished CDI-positive and -negative patients, with the greatest differentiation between the cohorts observed based on bacterial metatranscriptomics. Bipartite network analyses demonstrated that Aspergillus and Penicillium taxa shared a strong positive relationship in CDI patients and together formed negative co-occurring relationships with several bacterial taxa, including the Oscillospira, Comamonadaceae, Microbacteriaceae, and Cytophagaceae. The investigators identified new pathways associated with C. difficile infections. Those pathways pointed to still other bacterial species, including Escherichia coli that may contribute to the dysbiosis, or imbalance among bacterial species in the gut, associated with C. difficile.
David B. Stewart, MD, a surgeon and first author of the study, said, “The development of C. difficile infection is apparently influenced both by bacterial pathogens and fungi particular to this disease. Until now, fungi have been understudied and under-appreciated in the gut microbiome.” The study was published on August 28, 2019, in the journal mSphere.
Related Links:
University of Arizona