Gene Deletions Affect Rapid Diagnostic Tests for Malaria
By LabMedica International staff writers
Posted on 25 Jul 2019
Prompt and accurate diagnosis of malaria is crucial for malaria case-management and control and elimination programmes. While malaria diagnosis was historically based on symptoms alone, guidelines state that parasite-based diagnosis of malaria should be confirmed before treatment is given.Posted on 25 Jul 2019
Malaria rapid diagnostic tests (mRDT) that target histidine-rich protein 2 (HRP2) are important tools for Plasmodium falciparum diagnosis. Parasites with pfhrp2/3 gene deletions threaten the use of these mRDTs, and have been reported in Africa, Asia, and South America. The absence of both HRP2 and HRP3 renders the parasites undetectable by HRP2-based mRDTs.
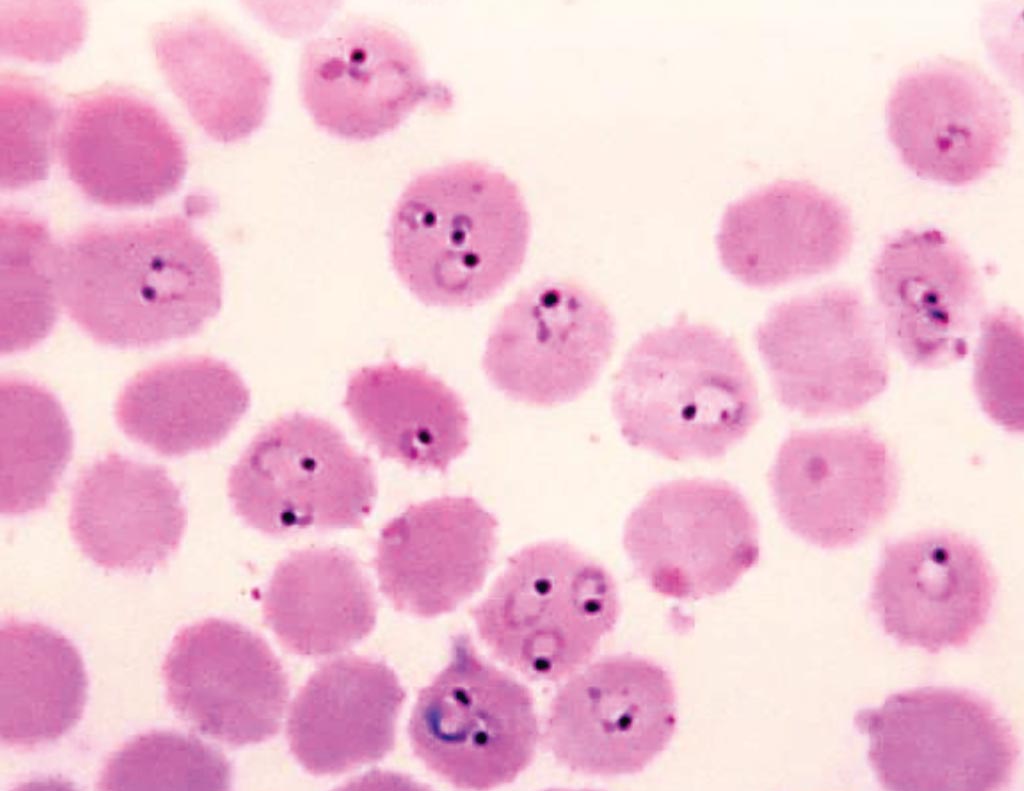
Image: A Giemsa-stained, thin film blood smear that shows the presence of numerous of ring-form, Plasmodium falciparum trophozoites, with some infected red blood cells (RBCs) harboring multiple organisms (Photo courtesy of Steve Glenn/CDC).
Scientists at the London School of Hygiene & Tropical Medicine (London, UK) and their colleagues analyzed P. falciparum parasites identified in human blood samples from three malaria studies in Ghana, Tanzania and Uganda. They analyzed 911 dried blood spots 165 from Ghana, 176 from Tanzania and 570 from Uganda (570). P. falciparum infection was confirmed by 18SrDNA polymerase-chain reaction (PCR), and pfhrp2/3 genes were genotyped.
True pfhrp2/3 gene deletions were confirmed if samples were (1) microscopy positive, (2) 18SrDNA polymerase chain (PCR) positive, (3) positive for merozoite surface protein genes by PCR, or positive by loop-mediated isothermal amplification (LAMP), and (4) quantitative PCR positive with > 5 parasites/µL. To confirm the deletion of pfhrp2 and pfhrp3 genes, PCR of two other single copy genes was performed.
The team reported that among the samples from Ghana, 107/165 (64.9%) were recorded as positive by mRDT and 82/165 (49.7%) by microscopy. In Tanzania, 72/171 (53.8%) samples were recorded as positive by mRDT, while 140/176 (79.6%) were positive by microscopy. Of the 570 Uganda samples, 258/570 (45.3%) were recorded as positive by mRDT, and 203 (35.6%) were positive by microscopy. No pfhrp2/3 deletions were detected in samples from Ghana, but deletions were identified in Tanzania (three pfhrp2; two pfhrp3) and Uganda (seven pfhrp2; two pfhrp3). Of the 10 samples with pfhrp2 deletions, nine tested negative by HRP2-based mRDT.
The authors concluded that the presence of pfhrp2/3 deletions in Tanzania and Uganda, along with reports of pfhrp2/3-deleted parasites in neighboring countries, reinforces the need for systematic surveillance to monitor the reliability of mRDTs in malaria-endemic countries. The study was published on June 28, 2019, in the Journal of Infectious Diseases.
Related Links:
London School of Hygiene & Tropical Medicine