Nasal Microbiome Influences Pneumonia and Related Diseases
By LabMedica International staff writers
Posted on 15 Jul 2019
A recent study examined the influence of the natural microbial flora in the nose and viral co-infection on the acquisition of Streptococcus pneumoniae bacteria and the development of pneumococcal diseases.Posted on 15 Jul 2019
S. pneumoniae is the main bacterial pathogen involved in pneumonia. It has been speculated that acquisition of the bacteria and colonization density was probably affected by viral co-infections, the local microbiome composition, and mucosal immunity. To examine the relationship between the nasal microbiome and acquisition of S. pneumoniae bacteria, investigators at Liverpool School of Tropical Medicine (United Kingdom) and the University of Edinburgh (United Kingdom) used an experimental human challenge model (the Experimental Human Pneumococcal Challenge) and inoculated live bacteria in combination with a live virus in the form of the readily available nasal vaccine for influenza.
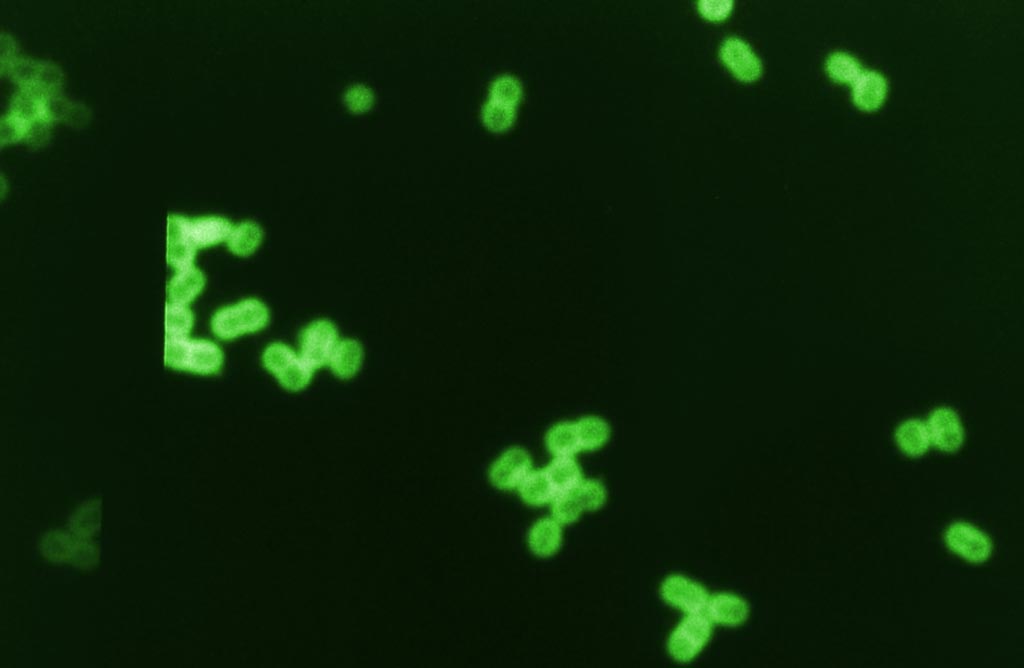
Image: Digitally colorized photomicrograph of fluorescent antibody labeled Streptococcus pneumoniae bacteria in spinal fluid sample (Photo courtesy of the CDC).
Working with this model, the investigators examined the interactions between live-attenuated influenza vaccine (LAIV), successive pneumococcal challenge, and the healthy adult nasal microbiota and mucosal immunity.
Results indicated that the equilibrium between the nasal microbiome and the host immune system had an impact on pneumococcal acquisition and density, in particular when combined with a viral co-infection. The microbial flora in the nose also appeared to have an effect on replication of the pathogenic virus itself.
Contributing author Dr. Daniela Ferreira, professor of respiratory vaccines and infection immunology at Liverpool School of Tropical Medicine, said, "We knew relatively little about the relationship between viral infections and the microbiota. Our model helped us to establish a link between baseline microbiota and colonization with the bacteria which causes pneumonia and shows the way that it is apparently altered with the introduction of a viral pathogen."
Senior author Dr. Debby Bogaert, professor of inflammation research at the University of Edinburgh, said, "Using this sophisticated human challenge model, we were for the first time able to identify that different constellations of microbes in the nose are associated with more or less inflammation, viral replication, and pneumococcal carriage receptiveness."
The work was published in the July 5, 2019, online edition of the journal Nature Communications.
Related Links:
Liverpool School of Tropical Medicine
University of Edinburgh