Rapid Assay Detects Pyrazinamide-Resistant Tuberculosis
By LabMedica International staff writers
Posted on 27 May 2019
Tuberculosis (TB), caused by Mycobacterium tuberculosis complex (MTBC) strains, remains one of the most frequent infectious diseases worldwide. Due to its high frequency and the increased risk of co-resistance to other drugs, rapid detection of pyrazinamide (PZA) resistance is essential to control the MDR-TB epidemic.Posted on 27 May 2019
Pyrazinamide is a key antibiotic for the treatment of drug susceptible tuberculosis. PZA-resistance is mainly mediated by mutations in the pncA gene; however the current gold standard is a phenotypic drug susceptibility test requiring a well-adjusted pH-value for reliable results. However, PZA resistance testing is not routinely performed.

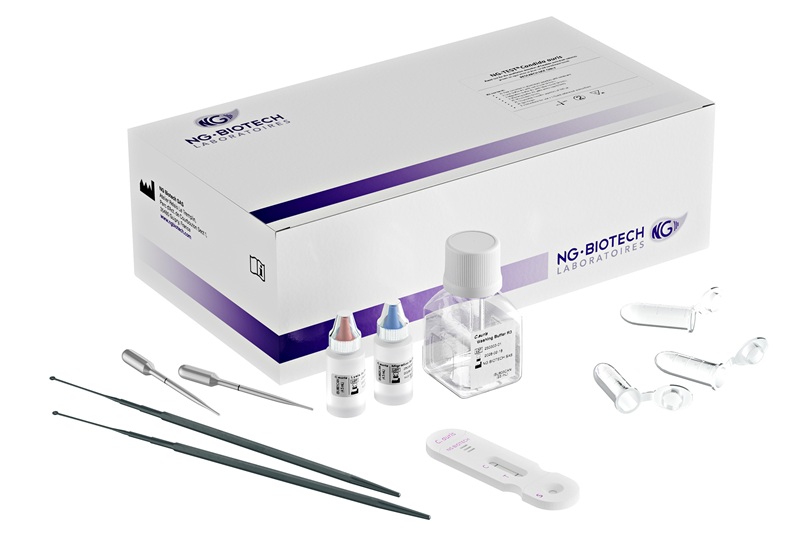
Image: The Alere q is a fully automated nucleic acid testing platform that brings the power of molecular testing to bench tops in any healthcare setting (Photo courtesy of Alere Technologies).
A team of microbiologists working with Alere Technologies GmbH (Jena, Germany) used 271 clinical MTBC isolates, which were derived from a national survey of drug resistance in Swaziland, for the clinical evaluation of the melting curve assay. All isolates were previously analyzed by Sanger sequencing of the pncA gene using flanking primers from position −104 to 560 out of 561 coding nucleotides. The sequence analysis showed wild type genotype for 153 isolates and different mutations in the analyzed pncA sequence for 118 isolates.
To test rare mutations (Ala25Ala, Ser74Ser, Leu116Arg and Asn147Asn) plasmids with the respective sequences were generated by Eurofins Genomics GmbH, (Ebersberg, Germany). The array probes had a C7-amino linker for immobilization on the solid array phase. The forward primers for the melting curve analysis were Cy5 labeled at the 5′ end, whereas the reverse primers for the melting curve analysis had no label as well as the primers for the TaqMan analysis. The amplification and melting curve assay was performed in a prototype cartridge produced by Alere Technologies GmbH.
The investigators reported that the melting curve assay detects a non-wild type genotype in selected pncA regions in at least 3,750 gene copies/mL within 2.5 hours. The performance of the melting curve assay was evaluated with 271 clinical isolates from Swaziland. Among 118 isolates with 21 different pncA mutations, 98 (83.05%) were detected as a non-wild type genotype. In comparison to Sanger sequencing, the melting curve assay showed a sensitivity of 83.05% and a specificity of 100%.
The authors concluded that the current prototype cartridge comprises 57 different probes but has a maximum capacity of 88 probes to be analyzed by microarray hybridization. The assay already detects reliably MTBC DNA down to 3,750 gene copies/mL with a turnaround time of 2.5 hours and works with genomic DNA as well as with cell lysates. The test can be automatically performed in a closed cartridge using the battery powered Alere q analyzer and has the potential to be applied in resource-limited settings as a point-of-care test allowing for a rapid inclusion of PZA into early treatment regimens. The study was published in the June 2019 issue of the journal Diagnostic Microbiology and Infectious Disease.