Gut Microbes Differ Between Two Common Conditions
By LabMedica International staff writers
Posted on 01 Jan 2019
Inflammatory bowel disease (IBD) and irritable bowel syndrome (IBS) are two of the most common diseases of the gastrointestinal tract. Unlike IBD, IBS does not cause inflammation, ulcers or other damage to the bowel.Posted on 01 Jan 2019
Instead, IBS is a much less serious problem called a functional disorder. This means that the digestive system looks normal but does not work as it should. Symptoms of IBS may include cramps, bloating, gas, diarrhea, constipation and mucus in the stool. IBS has also been called spastic colon or spastic bowel.
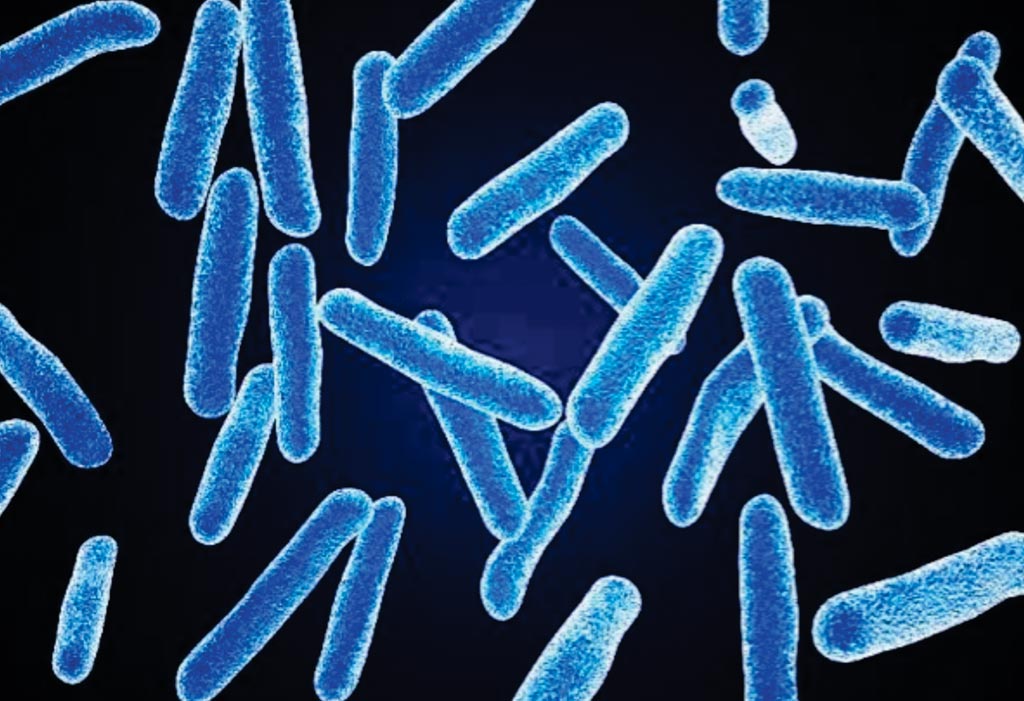
Image: A colored scanning electron micrograph (SEM) of Faecalibacterium prausnitzii, one of the most abundant anaerobic bacteria in the human gut microbiota, and its relative abundance is a biomarker of intestinal health in adults (Photo courtesy of BioFoundations).
An international team of scientists led by the University of Groningen (Groningen, Netherlands) performed metagenomic sequencing on stool samples from 355 individuals with IBD (Crohn's disease or ulcerative colitis), 412 individuals with IBS, and 1,025 unaffected, healthy controls, searching for microbial species and strains associated with one or both inflammatory gut conditions.
The team performed shotgun metagenomic sequencing used Illumina instruments on microbial DNA in home-collected stool samples from 1,792 well-phenotyped cases and controls from three cohorts from the Netherlands. The team's sequence data led to 219 Crohn's disease-related taxa, 102 taxa associated with ulcerative colitis, and 66 taxa with apparent ties to IBS. There was some overlap between conditions: Within IBD, for example, at least 87 of the microbes implicated in ulcerative colitis also turned up in individuals with Crohn's disease.
The scientist noted that the diversity of strains within beneficial bacterial species, such as Faecalibacterium prausnitzii, tended to wane in the guts of individuals with IBD or IBS relative to the unaffected controls. They also saw shifts in strain diversity for 21 bacterial species in individuals with Crohn's disease, compared to strain diversity changes in 15 species in those with ulcerative colitis and one species in the IBS group. The team also estimated growth rates for a subset of the gut microbial species, identifying small sets of species that seemed to have altered abundance in each of the conditions. In addition, they investigated the differences in bacterial composition, gene content, and gene functions in the IBD and IBS cases.
The authors concluded that despite substantial overlap between the gut microbiome of patients with IBD and IBS compared with control individuals, they were able to use gut microbiota composition differences to distinguish patients with IBD from those with IBS. By combining species-level profiles and strain-level profiles with bacterial growth rates, metabolic functions, antibiotic resistance, and virulence factor analyses, they identified key bacterial species that may be involved in two common gastrointestinal diseases. The study was published on December 19, 2018, in the journal Science Translational Medicine.
Related Links:
University of Groningen