Cell-Free DNA Sequencing Confirms Myelodysplastic Syndrome Diagnosis
By LabMedica International staff writers
Posted on 25 Aug 2016
The use of next-generation sequencing (NGS) methods to analyze cell-free DNA (cf-DNA) in the blood of patients with myelodysplastic syndrome (MDS) yields more accurate results than the current standard approach of Sanger sequencing.Posted on 25 Aug 2016
Myelodysplastic syndromes (MDSs) are a heterogeneous group of disorders, characterized by the presence of clonal neoplastic cells in bone marrow that lead to ineffective hematopoiesis and peripheral blood (PB) cytopenias. The major criteria for the diagnosis of MDS are the presence of peripheral cytopenia and dysplasia.

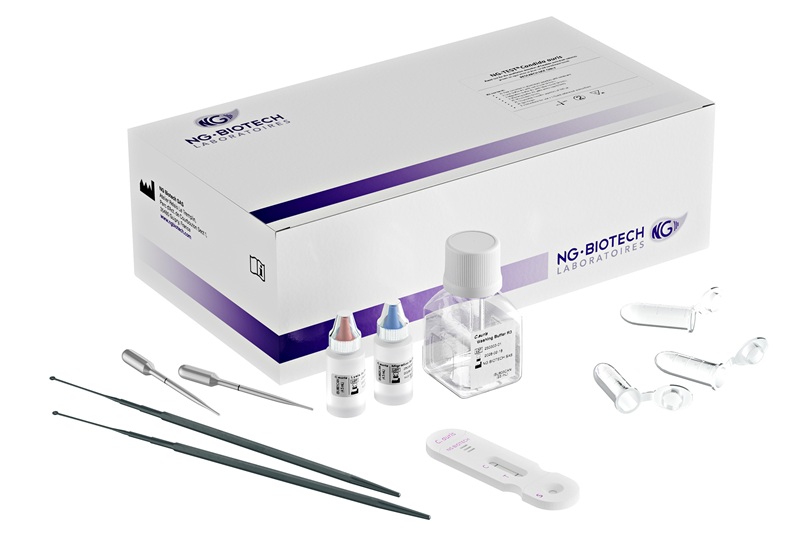
Image: The NucliSENS EasyMAG automated platform for total DNA extraction (Photo courtesy of BioMérieux).
Scientists at the NeoGenomics Laboratories (Irvine, CA, USA) collected PB samples were collected from 16 patients confirmed to have MDS by bone marrow evaluation and from four age-matched normal controls. All patients had blasts of less than 5%. This included patients with refractory cytopenia with unilineage dysplasia, refractory cytopenia with multilineage dysplasia, and refractory anemia with ring sideroblasts (RARS). The team extracted cf-DNA from plasma and cellular DNA from PB cells.
Total nucleic acid was isolated from plasma using NucliSENS EasyMAG automated platform (BioMérieux, Marcy-l'Étoile, France). DNA was then quantified using Qubit 2.0 Fluorometer (Thermo Fisher Scientific, Waltham, MA, USA). NGS and Sanger Sequencing were performed for several specific genes. Polymerase chain reaction products were purified and sequenced in both forward and reverse directions using an ABI PRISM 3730XL Genetic Analyzer (Applied Biosystems, Foster City, CA, USA).
Upon deep sequencing of the plasma cf-DNA, all 16 patients showed at least one mutated gene, confirming the presence of an abnormal clone consistent with MDS. No abnormality was detected in any of the four samples from normal control individuals. Eight patients (50%) showed mutation in one gene and the remaining eight patients (50%) showed mutations in two or more genes. Five of the 16 patients (31%) showed mutations detected by NGS of cf-DNA, while Sanger sequencing of PB cellular DNA showed no evidence of mutation.
Garth D. Ehrlich, PhD, FAAAS, Editor-in-Chief of the journal where the study was published, said, “This is a remarkable finding that cell-free DNA provides a more sensitive assay for detecting cancer than does the traditional approach of analyzing cell-associated DNA.” The study was published on July 14, 2016, in the journal Genetic Testing and Molecular Biomarkers.
Related Links:
NeoGenomics Laboratories
BioMérieux
Thermo Fisher Scientific
Applied Biosystems