Listeria Hypervirulence Uncovered by Harnessing Its Biodiversity
By LabMedica International staff writers
Posted on 14 Feb 2016
The Listeria monocytogenes bacterium responsible for the severe foodborne infection, listeriosis, especially in pregnant women and the elderly is subjected to a stringent microbiological monitoring system.Posted on 14 Feb 2016
Human epidemiological and clinical data with bacterial population genomics have been integrated to harness the biodiversity of the model foodborne pathogen L. monocytogenes and decipher the basis of its neural and placental tropisms.
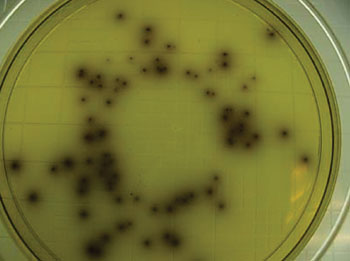
Image: Colonies of typical Listeria monocytogenes as they appear when grown on Listeria-selective agar (Photo courtesy of Dr. James Folsom).
Scientists at the Institute Pasteur (Paris, France) and their colleagues studied close to 7,000 strains of L. monocytogenes isolates collected over the past nine years for monitoring purposes. The bacterial molecular genotyping revealed considerable heterogeneity within the L. monocytogenes species, and showed that strains can be categorized into distinct genetic families (or clonal groups). By analyzing epidemiological data, the investigators showed that some of these clonal groups are more frequently associated with human infections, while others are closely linked to food.
The comprehensive analysis of detailed clinical data from over 800 patients with listeriosis showed that the strains most often associated with infections are more frequently isolated in the least immunodeficient patients while the strains most commonly linked to food mainly infect the most immunodeficient individuals. In addition, the strains most often involved in infections appear to be the most invasive as they infect the central nervous system and fetus more often than the strains most commonly associated with food.
To uncover the genetic basis of this hypervirulence, the teams sequenced the genomes of around a hundred strains that are representative of the most prevalent clonal groups. Comparative analysis of these genome sequences identified a large number of genes closely linked to the hypervirulent clonal groups, including one which was demonstrated as involved in the cerebral and fetal-placental tropism of L. monocytogenes experimentally. These results pave the way to a detailed understanding of the mechanisms underlying L. monocytogenes central nervous system and fetal-placental invasion. The study was published on February 1, 2016, in the journal Nature Genetics.
Related Links:
Institute Pasteur