Globally Accurate Diagnostic Test Developed for Herpes
By LabMedica International staff writers
Posted on 25 Aug 2015
A universally accurate diagnostic test for human herpes simplex viruses (HSV) may soon be developed and that may also lead to the development of a vaccine that protects against the virus.Posted on 25 Aug 2015
Currently, individuals are screened for HSV using a test that distinguishes between a glycoprotein, a molecule containing a carbohydrate and a protein present in HSV1, which is common throughout the population, and the considerably rarer HSV2. Whereas the test discriminates between the two variants with high accuracy in the USA and Europe, it largely fails in Africa, where rates of human immunodeficiency virus (HIV) and HSV are highest.
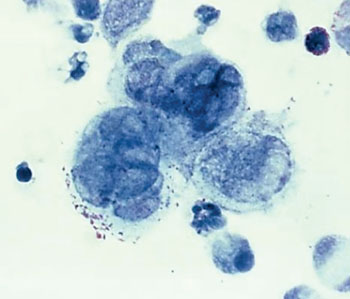
Image: Photomicrograph of the herpes simplex virus, within tissue taken from a penile lesion of a patient with genital herpes (Photo courtesy of the Centers of Disease Control and Prevention).
Scientists at National Institutes of Health (Bethesda, MD, USA) working with their academic colleagues and a team at Bioinfoexperts, LLC (Thibodaux, Louisiana, USA) used a variety of sequence analysis methods to compare all available sequence data for HSV-1 and HSV-2 glycoproteins, using viruses isolated in Europe, Asia, North America, the Republic of South Africa, and East Africa.
Published HSV-1 and HSV-2 glycoprotein sequences and their location data (when available) were downloaded from the Virus Pathogen Resource. Automated alignments were generated for each HSV species glycoprotein at the amino acid level and optimized by hand when necessary, after which the alignments were analyzed as nucleic acids. Alignments containing both HSV-1 and HSV-2 sequences for each glycoprotein were also generated.
The team reported that compared to HSV1, HSV2 has less genetic diversity. Besides providing clues to how the two strains evolved, the findings also have implications for vaccine development; because HSV2’s low genetic diversity means fewer antigens could be enough for developing a globally effective HSV2 vaccine. Because the current test discriminates between HSV1 and HSV2 by looking at variations in a localized region of so-called glycoprotein G, the team focused on the glycoproteins present in HSV by comparing the 36 HSV2 strains to 26 previously sequenced strains of HSV1 and looking at geographic diversity among the HSV2 glycoprotein sequences. They found that found that the African strains of glycoprotein G differed slightly from those in strains from other countries. Glycoproteins I and E also showed some variation.
Thomas C. Quinn, MD, a professor of medicine and coauthor of the study, said, “These variations explain why the diagnostic test didn’t work optimally in Africa. From this study, you then can make a consensus sequence that is common across the world for HSV2 glycoprotein that is different for HSV1 so you don’t get this misdiagnosis. It should now be possible to develop a more universal screening tool.” The studies were published in the August 2015 issue of the Journal of Virology.
Related Links:
National Institutes of Health
Bioinfoexperts, LLC