Efficient Molecular Method Detects Pathogenic Leptospires
By LabMedica International staff writers
Posted on 30 Jun 2015
The diagnosis of leptospirosis, which primarily relies on antiquated serotyping methods, is particularly challenging due to presentation of non-specific symptoms shared by other febrile illnesses, often leading to misdiagnosis. Posted on 30 Jun 2015
Pathogenic Leptospira species cause a prevalent yet neglected zoonotic disease with mild to life-threatening complications in a variety of susceptible animals and humans. Initiation of antimicrobial therapy during early infection to prevent more serious complications of disseminated infection is often not performed because of a lack of efficient diagnostic tests.

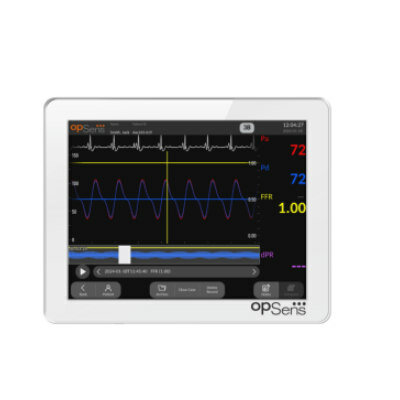
Image: The CFX96 real-time polymerase chain reaction (PCR) detection system (Photo courtesy of Bio-Rad Laboratories).
Scientists at the University of Maryland, College Park, MD, USA) and their colleagues collected blood samples from 25 patients enrolled during active surveillance for leptospirosis at a state-run hospital in Salvador (Brazil), from June 2013 to September 2013, and the samples were collected during early hospitalization. All patients were examined for leptospirosis using previously described methods hemoculture and microscopic agglutination test (MAT) as well as DNA quantitative polymerase chain reaction (qPCR). Confirmed patients were positive for at least one of the above three tests. It was determined that 22 out of 25 subjects had laboratory-confirmed leptospirosis.
Specific regions of the leptospiral 16S ribosomal ribonucleic acid (rRNA) molecules constituted a novel and efficient diagnostic target for PCR-based detection of pathogenic Leptospira serovars. All qPCR reactions were performed using the CFX96 real time PCR detection system (Bio-Rad Laboratories, Inc.; Hercules, CA, USA). The diagnostic test using spiked human blood was at least 100-fold more sensitive than corresponding leptospiral DNA-based quantitative PCR assays, targeting the same 16S nucleotide sequence in the RNA and DNA molecules. The sensitivity and specificity of the RNA assay against laboratory-confirmed human leptospirosis clinical samples were 64% and 100%, respectively, which were superior to an established parallel DNA detection assay.
The team discovered that 16S transcripts remain appreciably stable ex vivo, including untreated and stored human blood samples, further highlighting their use for clinical detection of L. interrogans. Together, these studies underscore a novel utility of RNA targets, specifically 16S rRNA, for development of PCR-based modalities for diagnosis of human leptospirosis, and also may serve as paradigm for detection of additional bacterial pathogens for which early diagnosis is warranted. The study was published on June 19, 2015, in the journal Public Library of Science ONE.
Related Links:
University of Maryland
Bio-Rad Laboratories