Nasal Bacteria Could Predict Skin Infections
By LabMedica International staff writers
Posted on 05 Jun 2014
Bacteria found in the nose may be a key indicator for future development of skin and soft-tissue infections in remote areas of the body. Posted on 05 Jun 2014
The nose is the primary reservoir of Staphylococcus aureus in humans and in nearly 80% of the cases, an individual's colonizing strain is the same strain that causes subsequent remote skin infections. However, until now, no one has been able to determine why some S. aureus carriers develop infections while others do not.
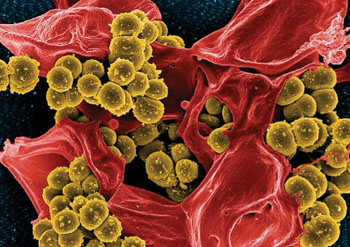
Image: Scanning electron micrograph of Methicillin-resistant Staphylococcus aureus and a dead human neutrophil (Photo courtesy of the US National Institute of Allergy and Infectious Diseases).
Scientist at the Uniformed Services University of the Health Sciences (Bethesda, MD, USA) postulated that the population of S. aureus in an individual's nose may harbor valuable clues regarding skin and soft tissue infections (SSTI) susceptibility that had not yet been described. This is of particular interest to the military, as it is well known that soldiers in training are at increased risk of developing an SSTI. They collected nasal samples and cultures from 86 infantry soldiers and for those individuals among the 86 who developed SSTIs, the investigators also collected samples and cultures from within the soldiers' skin abscesses.
The team used a high-throughput DNA sequencing strategy to determine the microbial composition of each sample. The biodiversity of the bacterial population in each nose was compared between individuals colonized and/or infected with Methicillin-Resistant S. aureus (MRSA), Methicillin-Sensitive S. aureus (MSSA), and those individuals that were culture-negative for S. aureus.
The scientists observed a significantly higher percentage of a type of bacteria known as Proteobacteria in the noses of individuals who did not develop SSTI, suggesting that Proteobacteria may, in fact, be protective against the development of SSTIs. Furthermore, S. aureus carriers had a unique nasal microbiome that differed from non-carriers. The Proteobacteria are a major phylum of bacteria. They include a wide variety of pathogens, such as Escherichia, Salmonella, Vibrio, Helicobacter, and many other notable genera.
The scientists concluded that by establishing a nose marker microbiome associated with development of SSTI infections may pave the way for focused preventive treatments that target the microbiome, rather than S. aureus itself. They believe that this study will aid in the design of future prophylactic procedures that can help prevent SSTI, particularly in the setting of military training, and help influence how health care providers think about and treat these complex and diverse infections. The study was presented at the 114th General Meeting of the American Society for Microbiology held May 17–20, 2014, in Boston (MA, USA).
Related Links:
Uniformed Services University of the Health Sciences