Novel Subtyping Assay Detects Clostridium Virulence Genes
By LabMedica International staff writers
Posted on 29 Jan 2014
The application of a novel nucleic acid detection platform to detect Clostridium difficile in subjects presenting with acute diarrheal symptoms has been demonstrated. Posted on 29 Jan 2014
Current methods of diagnosing C. difficile include stool culture, toxin testing, enzyme immunoassays, and polymerase chain reaction (PCR). However, these methods are impractical in most clinical settings, because they require two to three days to complete, during which time clinicians must rely on empirical treatment of disease with antibiotics.
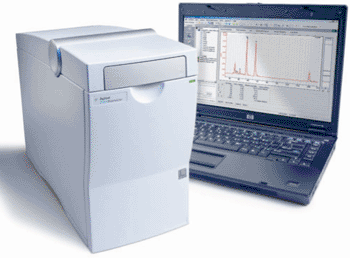
Image: The Agilent 2100 Bioanalyzer for DNA, RNA, and protein analysis (Photo courtesy of Agilent Technologies).
Scientists at Brown University (Providence, RI, USA) have developed a novel PCR assay coupled with a small-volume, real-time platform allowing for simple and rapid detection of three C. difficile genes: Clostridium difficile toxin B (tcdB), and tcdC, and cytolethal distending toxins binding (cdtB). Amplification of DNA from the tcdB, tcdC, and cdtB genes was performed using a droplet-based sandwich platform with quantitative real-time PCR (qPCR) in microliter droplets to detect and identify the amplified fragments of DNA. The product size was determined using Agilent DNA 1000 chips on an Agilent 2100 Bioanalyzer system (Agilent Technologies; Santa Clara, CA, USA).
The investigators identified the presence of C. difficile in clinical stool specimens through a series of three steps: isolation of double-stranded DNA, amplification of segments of DNA specific to C. difficile in genes of interest that may produce proteins conferring hyper-virulence, and detection of those PCR products through the use of qPCR or capillary electrophoresis. Three sets of PCR primers were designed to amplify three specific regions of DNA, each located within a gene with a potential role in coding for the production of proteins involved in the severity of illness associated with C. difficile infection.
The team concluded that their technique of multiplex gene amplification provides a unique method that is both sensitive and specific for rapidly detecting C. difficile in patient stool samples. This method can be adapted to point-of-care testing and thus can assist physicians in developing and implementing better treatment regimens for the care of patients with C. difficile infections, particularly those with the NAP1/027/BI strain. The study was published on January 13, 2014, in the Journal of Molecular Diagnostics.
Related Links:
Brown University
Agilent Technologies