Bacterial Isolates Identified by Colorimetric Cards
By LabMedica International staff writers
Posted on 21 Dec 2010
A rapid and accurate bacterial identification system for Staphylococci species has been validated in a clinical setting. Posted on 21 Dec 2010
A procedure that uses colorimetric identification card and automation has been compared with molecular methods to identify bacteria from patient's samples that were isolated and recovered during routine testing.
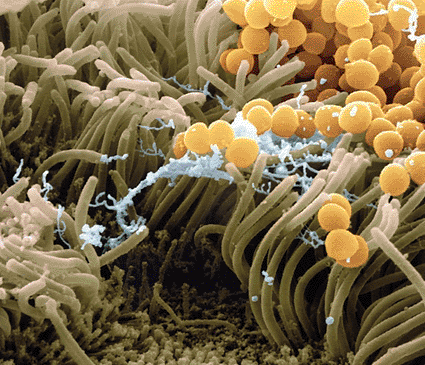
Image: Colored scanning electron micrograph (SEM) of Staphylococcus aureus bacteria (yellow) on human nasal epithelial cells (photo courtesy Juergen Berger / Science Photo Library).
Scientists from the Medical School at the University of Athens (Greece), isolated 147 clinical isolates: 52 Staphylococcus aureus, 50 Staphylococcus epidermidis, and 45 other coagulase-negative Staphylococci (CoNS). These isolates were recovered from various clinical samples including blood, wound, catheter tips, respiratory, and urine during routine testing, and for the majority of them, their isolation was considered clinically significant. The bacteria were first identified by molecular methodology and then comparatively tested by Vitek 2, colorimetric identification card, and Phoenix using the novel 0.25 McFarland and the standard 0.50 McFarland inoculum protocols.
All S. aureus isolates were accurately identified. Vitek 2 (bioMérieux, Marcy l'Etoile, France) identified correctly all S. epidermidis and 93.3% of the other CoNS, whereas the respective rates were 86% and 82.2% for Phoenix's standard and 92% and 82.2% for the novel protocol. The PMIC/ID-53 panel for Gram-positive cocci and software version V5.10A/V4.23D were used for identification by Phoenix system (Becton Dickinson Diagnostic Systems, Franklin Lane, NJ, USA).
The authors concluded that both systems provide excellent identification of S. aureus, but Vitek 2 recognizes CoNS species more accurately than Phoenix. The 0.25 McFarland protocol does not improve system performance. However, they also note specificity of testing is an important parameter and needs to be determined also. It is imperative that such evaluation studies use reference phenotypic or molecular identification methods for comparison. The study was published online on November 19, 2010 in Diagnostic Microbiology and Infectious Disease.
Related Links:
University of Athens
bioMérieux
Becton Dickinson Diagnostic Systems