T Cells Recognize Recent SARS-CoV-2 Variants
By LabMedica International staff writers
Posted on 15 Apr 2021
Due to the proofreading ability of the coronavirus (CoV) RNA-dependent RNA polymerase, the evolution of the global SARS-CoV-2 viral population during the current pandemic has been relatively constrained as compared to other endemic RNA viruses that do not possess this ability.Posted on 15 Apr 2021
During late 2020, three distinct variants that each possessed a significantly increased amount of amino acid polymorphisms were identified in association with spikes in cases of COVID-19 in the United Kingdom (variant B.1.1.7), South Africa (variant B.1.351), and Brazil (variant B.1.1.248). These variants all possess the N501Y mutation in the receptor-binding domain (RBD) of the SARS-CoV-2 spike protein, a primary target for neutralizing antibody (NAb) binding.
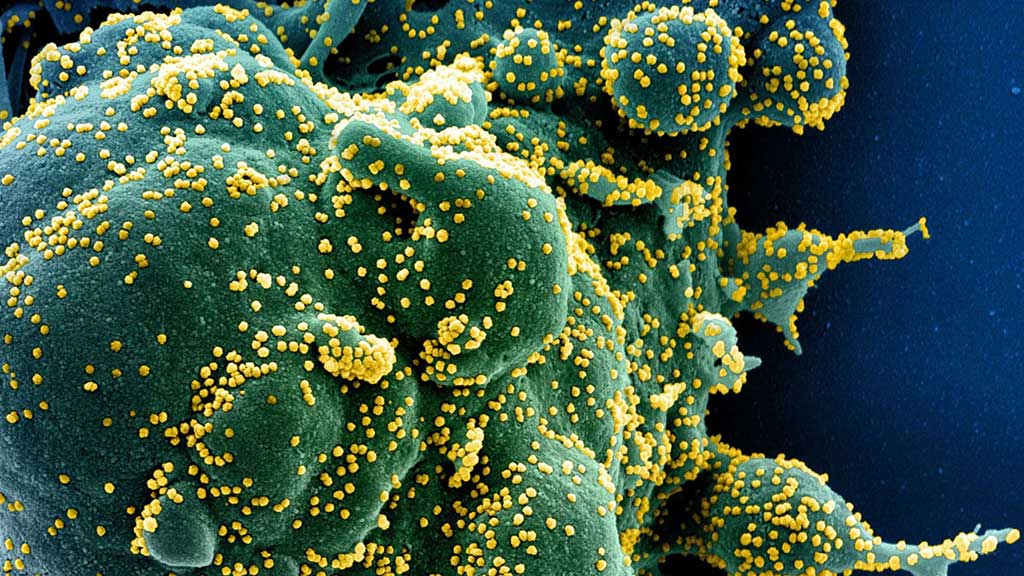
Image: Immune T cells lymphocytes can seek and destroy a cell (green) infected with and making copies of SARS-CoV-2 (yellow) (Photo courtesy of US National Institute of Allergy and Infectious Diseases)
A collaborating team of scientists led by the Johns Hopkins University School of Medicine (Baltimore, MD, USA) analyzed blood cell samples from 30 people who had contracted and recovered from coronavirus disease 2019 (COVID-19) prior to the emergence of virus variants. Most (60%) individuals included in the analysis were male and samples were collected a median of 42.5 days (interquartile range, 37.5-48 days) from initial diagnosis. The team aimed to determine whether CD8+ T cells in the blood could still recognize the three main SARS-CoV-2 variants. Peripheral blood mononuclear cell (PBMC) samples from the patients were collected and examined across six different human leukocyte antigen (HLA) haplotypes (HLAA*01:01, HLA-A*02:01, HLA-A03:01, HLA-A*11:01, HLA-A*24:02 and HLA-B*07:02).
A multiplexed peptide-MHC tetramer staining approach permitted the screening of 408 potential SARS-CoV-2 candidate epitopes for CD8+ T cell recognition. T cells were also evaluated using a 28-marker phenotypic panel (Immunoscape Pte. Ltd, Singapore). A total of 52 unique epitope responses were found and were directed against several structural and non-structural viral proteins. For controls, CD8+ T cells were probed for reactivity for up to 20 different SARS-CoV-2-unrelated control peptides per HLA (Adenovirus-, CMV-, EBV-, Influenza-, and MART-1-derived epitopes).
The team reported that only one mutation found in the B.1.351-Spike overlapped with a previously identified epitope (1/52), suggesting that virtually all anti-SARS-CoV-2 CD8+ T-cell responses should recognize these newly described variants. This mutation is the D80A mutation in the Spike protein, and occurs in the third residue of the RFDN VLPF epitope. This is a HLA*A24:02-restricted epitope for which a CD8+ T cell response was detected in 1/5 HLA*A24:02+ individuals, and at a low frequency (0.005 of total CD8+ T-cells), indicating this is not a high-prevalence epitope.
The authors concluded that their data highlighted the potential significant role of a multi-epitope T cell response in limiting viral escape, and partly mediate protection from disease caused by the SARS-CoV-2 variants. It is important that vaccines used for widespread campaigns generate strong multivalent T-cell responses in addition to neutralizing antibody (Nab) and other humoral responses in order to optimize efficacy against the current SARS-CoV-2 and emerging strains. It will be important to continue to monitor the breadth, magnitude, and durability of the anti-SARS-CoV-2 T cell responses in recovered and vaccinated individuals as part of any assessment to determine if booster vaccinations are needed. The study was published on March 30, 2021 in the journal Open Forum Infectious Diseases.
Related Links:
Johns Hopkins University School of Medicine
Immunoscape Pte. Ltd