Molecular Differences Discovered Among Ovarian Cancer Subgroups
By LabMedica International staff writers
Posted on 28 Apr 2020
Ovarian cancer is a highly diverse disease with a high rate of overall mortality. High-grade serous ovarian cancer (HGSC) is the most common and aggressive type of epithelial ovarian cancer, exhibiting high levels of tumor heterogeneity and variable clinical outcomes.Posted on 28 Apr 2020
The extent of residual disease following upfront cytoreductive surgery for HGSC is one of the strongest prognostic factors for progression-free and overall survival. Neoadjuvant chemotherapy (NACT) followed by interval cytoreductive surgery has been offered as an alternative approach, especially where full cytoreductive surgery is not feasible.
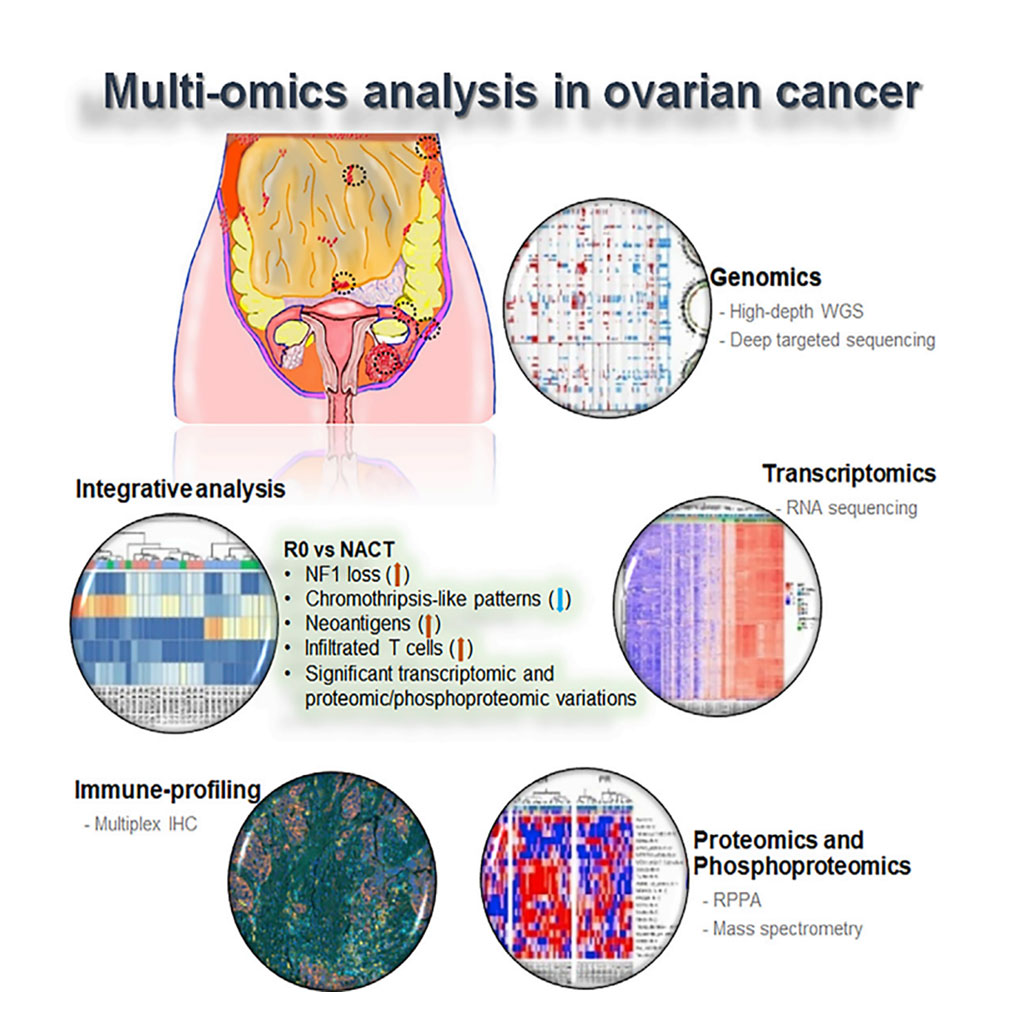
Image: Schematic Diagram of the Molecular Analysis of Clinically Defined Subsets of High-Grade Serous Ovarian Cancer (Photo courtesy of MD Anderson Cancer Center).
A team of scientists led by the University of Texas MD Anderson Cancer Center (Houston, TX, USA) analyzed tumor biopsies from 30 women with HGSC, some of whom had no visual evidence of disease after surgery, some of whom responded well to neoadjuvant chemotherapy, and some of whom responded poorly to neoadjuvant chemotherapy. This analysis indicated that tumors that underwent full resection differed molecularly from the other subgroups and that some of these differences between cancer subgroups could be applied to predict disease response.
The investigators collected primary tumor tissue and tissue from metastatic sites, which they analyzed using whole-genome sequencing to an average somatic coverage of 118× and to germline coverage of 38×. They also conducted targeted deep-sequencing, RNA sequencing, and proteomic and immune profiling on these samples. This analysis indicated that tumors that underwent full resection differed molecularly from the other subgroups and that some of these differences between cancer subgroups could be applied to predict disease response.
The scientists reported that across all both primary and metastatic samples, the most frequently mutated gene was TP53. However, the types of mutations affecting TP53 varied by tumor subgroup. For instance, the subgroups that underwent neoadjuvant chemotherapy (NACT) were more likely to have nonsense TP53 mutations, while the complete gross resection (R0) group had missense mutations. Other alterations varied by subgroup. About 54% of R0 group tumors had copy number loss of NF1, while the NACT groups had much lower levels of NF1 loss, about 18%. NF1 mRNA and protein levels were similarly decreased in the R0 group as compared to the NACT groups. NF1, they noted, could be a marker to predict R0 and NACT response.
R0 subgroup tumors additionally exhibited a strong binding neoantigen signature, as compared to the NACT subgroups. This neoantigen signature, they noted, is associated with the increased infiltration of tumor immune cells. When they examined the immune profiles of the tumors, the researchers found the R0 group exhibited an increased number of infiltrated T cells, as compared to the NACT subgroups. The NACT subgroups, by contrast, had increased macrophages and B cells, as compared to the R0 group. Meanwhile, R0 subgroup tumors also had lower levels of chromothripsis than the other subgroups. Further, when chromothripsis did occur in those tumors, it was more likely to affect chromosomes 6 and 19. Chromothripsis in NACT subgroup tumors was, by contrast, more likely to affect chromosomes 8 and 17.
The authors concluded that they had identified significant distinct molecular abnormalities and cellular changes and immune cell repertoire alterations between the groups, including a higher rate of NF1 copy number loss, and reduced chromothripsis-like patterns, higher levels of strong-binding neoantigens, and a higher number of infiltrated T cells in the R0 versus the NACT groups. The study was published on April 14, 2020 in the journal Cell Reports.
Related Links:
University of Texas MD Anderson Cancer Center