Epigenomic Biomarker Predict CAR T-Cell Therapy Resistance in Pediatric Leukemia
Posted on 18 Apr 2022
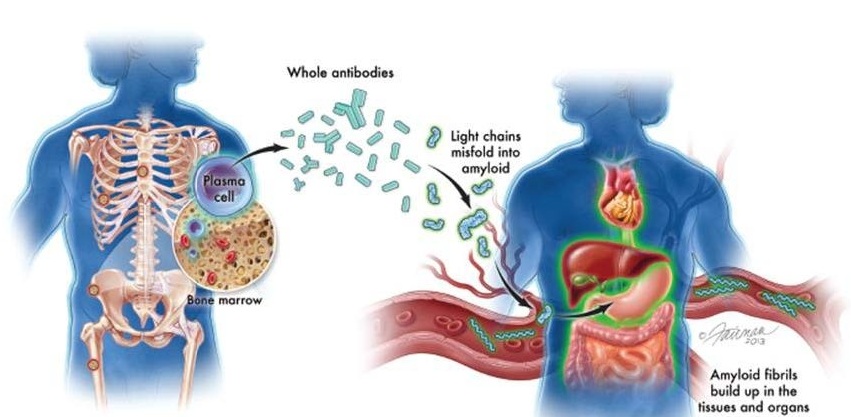
Children with acute lymphoblastic leukemia (ALL) whose cells express an epigenomic biomarker rooted in DNA methylation patterns may be less likely to respond to CD19-directed CAR T-cell therapy.
CAR T-cell therapy is a type of immunotherapy in which immune cells called T cells are harvested from a patient, reprogrammed to target cancer cells, and infused back into the patient to fight the cancer. A common CAR T-cell target is the receptor CD19; however, cancer cells can mutate CD19 or suppress its expression to develop CAR T resistance. While CD19 expression is currently one of the few biomarkers of potential CAR T response, not all patients who develop resistance lose CD19 expression.

Medical Scientists collaborating with the Seattle Children's Hospital (Seattle, WA, USA) acquired pretreatment bone marrow samples from the PLAT-02 clinical trial. The samples were collected from seven patients who experienced a complete remission following CD19-targeted CAR T therapy and seven patients who did not have a response, defined by continued evidence of circulating leukemia cells 63 days post-treatment. The team performed extensive studies, including whole-exome sequencing, bulk RNA-sequencing, long-read sequencing of the CD19 locus, array-based methylation testing, ATAC sequencing, single-cell RNA sequencing, and CyTOF mass cytometry (Fluidigm, South San Francisco, CA, USA).
The investigators reported that they found 238 regions of increased DNA methylation in patients who did not respond to treatment, and further determined that the pre-treatment methylation patterns were those known to be turned off by polycomb repressive complex 2 (PRC2) repression in stem cells. They then did a gene set enrichment analysis of their ATAC sequencing data and saw increased accessibility of chromatin at regions known to be associated with proliferation and cell cycling in stem cells.
The team also identified decreased expression of genes involved in antigen presentation and processing, pathways that are crucial for mounting an immune response, in cells that did not respond to CAR T therapy. Decreased antigen presentation could indicate that, even if leukemia cells continue to express CD19, they may not effectively process additional immune targets.
Javed Khan, MD, the senior author of the study said, “Interestingly, we saw subpopulations of cells expressing both lymphoid and myeloid markers, indicating that the epigenomes of some nonresponsive leukemias may contain a hybrid population of cells with a hybrid of ALL and AML epigenomes. Our data suggest that these leukemias, characterized by both lymphoid and myeloid-specific accessible regions, are likely less differentiated than responsive leukemia.” The study was presented at the American Association of Cancer Research's annual meeting held April 8-13, 2022, in New Orleans, LA, USA.
Related Links:
Seattle Children's Hospital
Fluidigm