Molecular and Cytogenetic Characterization of Myelodysplastic Syndromes in Cell-Free DNA
Posted on 04 Mar 2022
Molecular and cytogenetic studies are essential in patients with myelodysplastic syndromes (MDS) for diagnosis and prognosis. Cell-free DNA (cfDNA) analysis has been reported as a reliable non-invasive approach for detecting molecular abnormalities in MDS, however, there is limited information about cytogenetic alterations and monitoring in cfDNA.
Myelodysplastic syndromes (MDS) are hematopoietic stem cell disorders characterized by dysplasia and ineffective hematopoiesis that are driven by somatically acquired genomic alterations.
Molecular studies and conventional cytogenetics are essential in MDS to establish a correct diagnosis and to set up accurate risk stratification. Routinely, these analyses are performed in bone marrow (BM) samples, in particular cytogenetic analysis as it is difficult to obtain metaphases from peripheral blood (PB) samples.
Clinical Scientists at the Institut Hospital del Mar d'Investigacions Mèdiques (Barcelona, Spain) and their colleagues assessed the molecular and cytogenetic profile of a cohort of 70 patients with MDS by next-generation sequencing (NGS) using cfDNA and compared the results to paired bone marrow (BM) DNA.
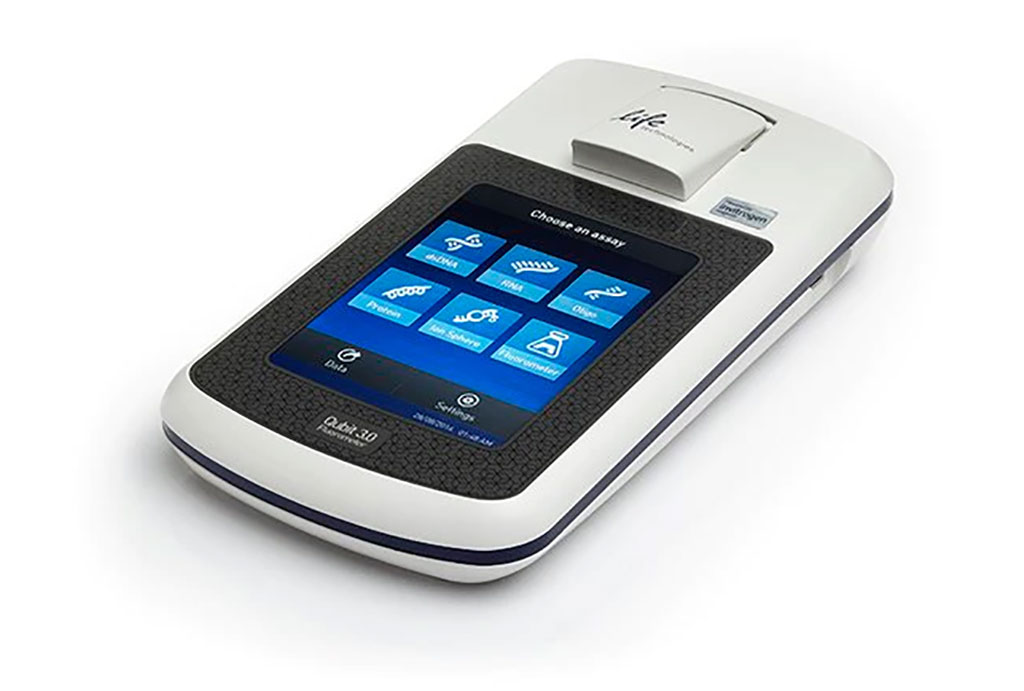
BM aspirates were collected and BM DNA was extracted with MagAttract DNA Blood Mini M48 Kit (Qiagen, Hilden, Germany). Cell-free DNA (CfDNA) was isolated automatically using Qiagen’s QIAsymphony SP (QIAsymphony DSP Virus/Pathogen Kit) and quantified with Qubit 3.0 (Thermo Fisher Scientific, Eugene, OR, USA). Genomic characterization was performed in paired sample of BM DNA and cfDNA by next-generation sequencing (NGS) in all patients. Libraries were prepared using a custom panel including 48 myeloid-associated genes. Libraries were sequenced with a 3000× minimum read depth in MiSeq/NextSeq (Illumina, San Diego, CA, USA).
The scientists reported that the amount of total cfDNA obtained in MDS patients (median: 58.4 ng/L) was significantly higher than that obtained from healthy controls (median: 32.4 ng/mL). A positive correlation was observed between the amount of cfDNA and the serum lactate dehydrogenase (LDH) levels. Mutational profiling of BM DNA and cfDNA showed comparable results: mutations were detected in BM DNA and cfDNA, with a 92.1% concordance.
The most frequently 140 mutated genes were TET2 (45.7%), SF3B1 (37.1%), ASXL1 (21.4%), DNMT3A (20.0%), SRSF2 141 (15.7%), ZRSR2 (11.4%) and U2AF1 (11.4%). A strong correlation was observed between the variant allele frequencies (VAF) of BM and cfDNA. The team compared the VAF of the detected mutations in cfDNA and BM DNA grouped by gene and observed that VAFs of SF3B1 mutations were significantly higher in cfDNA than in BM DNA. Cytogenetic/FISH alterations were detected at the time of diagnosis in 20/70 (28.6%) MDS patients. NGS analysis detected abnormalities in 10/70 MDS patients, in both BM DNA and cfDNA.
The authors concluded that that cfDNA mirrors the molecular profile of BM in MDS. In their cohort, enriched with lower risk patients, cytogenetic alterations were detectable in most cases by NGS in both BM DNA and cfDNA. Although further studies with larger cohorts are required to confirm these results, especially for cytogenetic alterations, our data support that the analysis of cfDNA is a promising method to characterize and monitor the molecular abnormalities present in patients with MDS. The study was published on February 22, 2022 in the journal Blood Advances.
Related Links:
Institut Hospital del Mar d'Investigacions Mèdiques
Qiagen
Thermo Fisher Scientific
Illumina