Checkpoint Protein Expression in Tumor Microenvironment Defines Prognosis of Hodgkin Lymphoma Patients
By LabMedica International staff writers
Posted on 04 Jan 2022
In Hodgkin lymphoma, B-lymphocytes start to multiply in an abnormal way and begin to collect in certain parts of the lymphatic system, such as the lymph nodes (glands). The affected lymphocytes lose their infection-fighting properties, making one more vulnerable to infection.Posted on 04 Jan 2022
In classical Hodgkin lymphoma (cHL), sparse malignant Hodgkin Reed-Sternberg (HRS) cells are embedded into extensive tumor microenvironment (TME) consisting mostly of benign immune cells, such as T and B lymphocytes, macrophages, eosinophils, plasma cells, and mast cells.
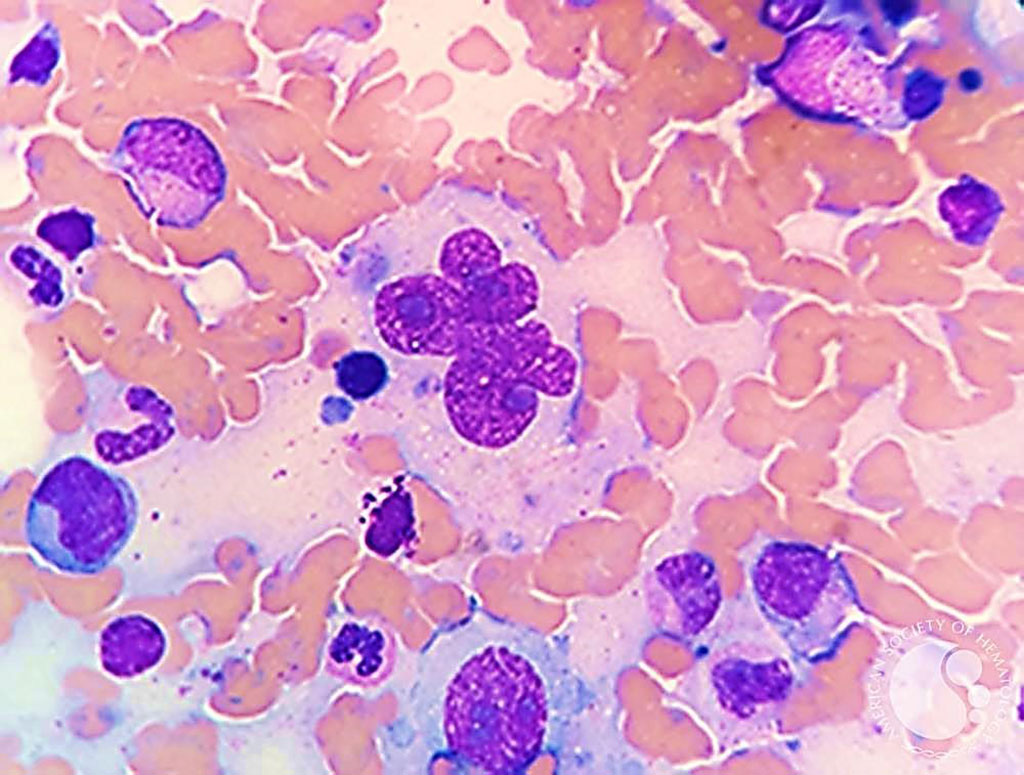
Image: Bone marrow aspirate from a patient with classical Hodgkin Lymphoma with large multinucleated Reed-Sternberg cells. “Hodgkin cells” are mononuclear while “Reed-Sternberg” cells are multinucleate forms (Photo courtesy of Nidia P. Zapata, MD and Espinoza-Zamora Ramiro)
Clinical Scientists at the Helsinki University Hospital (Helsinki, Finland) and their colleagues studied clinical data and formalin-fixed paraffin-embedded (FFPE) diagnostic tumor tissue samples from patients with primary cHL diagnosed between years 1993- 2012 and treated or followed. The patients were divided into two cohorts: the first cohort, “gene expression cohort”, consisted of 88 patients, and the second cohort, “immunohistochemistry (IHC) cohort”, of 131 patients.
The scientists used the NanoString nCounter digital gene expression profiling with 770-gene PanCancer Immune Profiling Panel (XT-CSO-HIP1-12, NanoString Technologies, Seattle, WA, USA) for gene expression profiling. To assess the proportions of the distinct immune cells in the TME, they applied CIBERSORT (Stanford University, Stanford, CA, USA) for the gene expression cohort, and multiplex immunohistochemistry to characterize the immunoprofile of cHL TME, and correlated the findings with survival. Gene expression analysis divided tumors into subgroups with T cell-inflamed and non-inflamed TME. Several macrophage-related genes were upregulated in samples with the non-T cell-inflamed TME, and based on the immune cell proportions, the samples clustered according to the content of T cells and macrophages.
The investigators reported that a cluster with high proportions of checkpoint protein (PD-1, PD-L1, IDO-1, LAG-3, and TIM-3) positive immune cells translated to unfavorable overall survival (OS) (5-year OS 76% vs. 96%), and remained as an independent prognostic factor for OS in multivariable analysis (HR 4.34). cHLs with high proportions of checkpoint proteins overexpressed genes coding for cytolytic factors, proposing paradoxically that they were immunologically active. This checkpoint molecule gene signature translated to inferior survival in a validation cohort of 290 diagnostic cHL samples and in an expansion cohort of 84 cHL relapse samples.
The authors concluded that their findings provide novel, more accurate information on the composition of different immune cells, checkpoint molecules, and their relationship in the heterogeneous cHL TME. Furthermore, the data recognize the prognostic impact of T cell and Tumor-Associated Macrophage (TAM) mediated checkpoint molecules on the survival of cHL patients. The study was published on December 23 2021 in the journal Blood Advances.
Related Links:
Helsinki University Hospital
NanoString Technologies
CIBERSORT