Targeted Locus Amplification Validated for Lymphoma
By LabMedica International staff writers
Posted on 15 Jul 2021
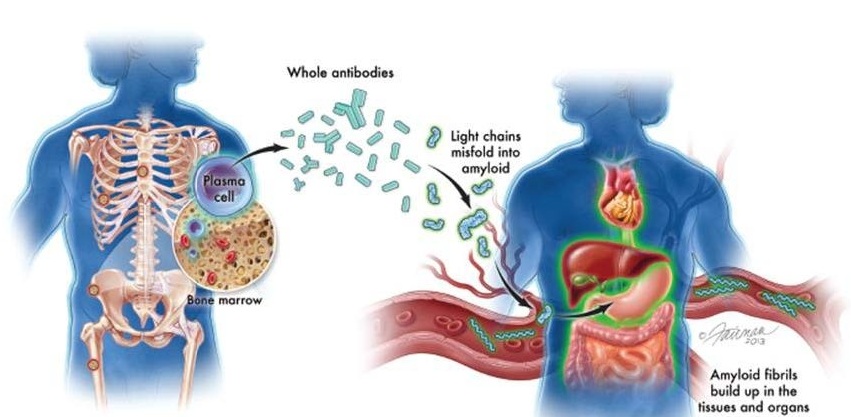
Structural variation (SV) in the genome is a recurring hallmark of cancer. Translocations (genomic rearrangements between chromosomes) in particular are found as recurrent drivers in many types of hematolymphoid malignancies. Posted on 15 Jul 2021
In routine diagnostic pathology, cancer biopsies are preserved by formalin-fixed, paraffin-embedding (FFPE) procedures for examination of intra-cellular morphology. Such procedures inadvertently induce DNA fragmentation, which compromises sequencing-based analyses of chromosomal rearrangements.
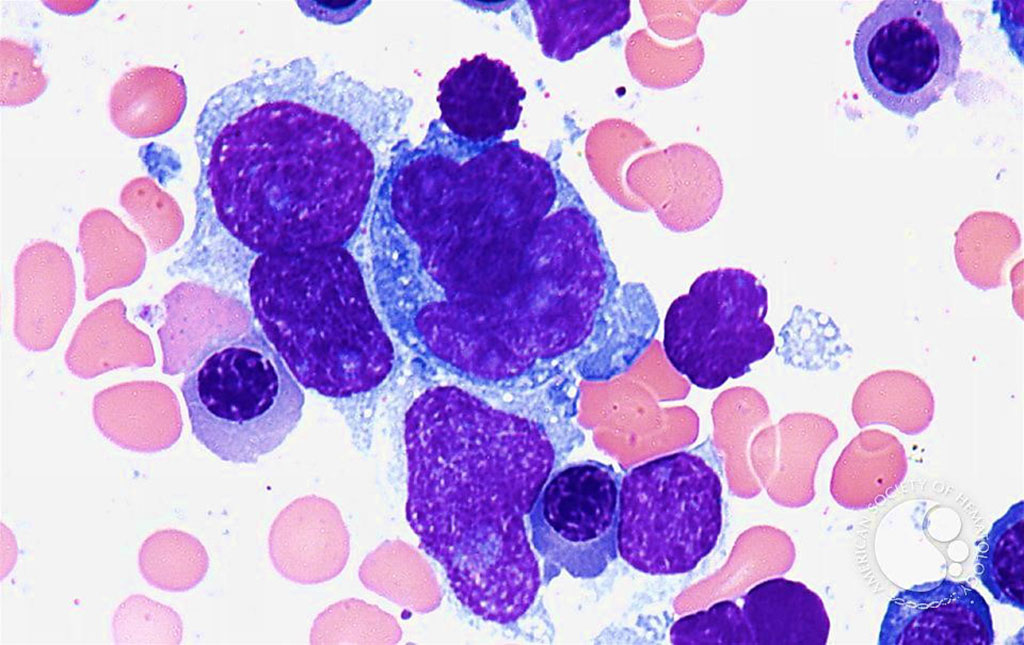
Image: Bone marrow aspirate from a patient with diffuse large B-cell lymphoma (Photo courtesy of Dr. Peter G. Maslak, MD)
Medical Scientists at the University Medical Center Utrecht (Utrecht, the Netherlands) and colleagues carried out a retrospective study using a set of 129 archival B-cell Non-Hodgkin lymphoma tissue samples, which were selected by the respective sites. The patients had been mostly diagnosed as Diffuse Large B-Cell Lymphoma (DLBCL), but also Burkitt, follicular and marginal zone lymphomas and some other diagnoses were included. Non-lymphoma control samples were also analyzed, mostly reactive lymph node samples and tonsillectomy specimens. FFPE tissue samples were obtained using standard diagnostic procedures.
The team developed FFPE-targeted locus capture (FFPE-TLC) method for targeted sequencing of proximity-ligation products formed in FFPE tissue blocks, and PLIER, a computational framework that allows automated identification and characterization of rearrangements involving selected, clinically relevant, loci. FFPE-TLC, blindly applied to 149 lymphoma and control FFPE samples, identifies the known and previously uncharacterized rearrangement partners. This method outperforms fluorescence in situ hybridization (FISH) in sensitivity and specificity, and shows clear advantages over standard capture-NGS methods, finding rearrangements involving repetitive sequences which they typically miss. The team included core needle biopsy samples in this study, which showed that even very small samples yielded good quality FFPE-TLC results.
The authors concluded that FFPE-TLC combined with PLIER for objective rearrangement calling offers clear advantages over regular NGS-capture approaches and over FISH for the molecular diagnosis of lymphoma FFPE specimens. The study was published on June 7, 2021, in the journal Nature Communications.
Related Links:
University Medical Center Utrecht