Single-Cell Sequencing Reveals Clonal Diversity Among AML Patients
By LabMedica International staff writers
Posted on 03 Nov 2020
A growing body of evidence supports the role of clonal diversity in therapeutic resistance, recurrence, and poor outcomes in cancer. Clonal diversity also reflects the history of the accumulation of somatic mutations within a tumor.Posted on 03 Nov 2020
The ability to infer clonal heterogeneity and tumor phylogeny from bulk sequencing data is inherently limited, because bulk sequencing techniques cannot reliably infer mutation co-occurrences and hence often fail in accurately reconstructing clonal substructure. Single-cell DNA sequencing (scDNA-seq) can address some of these challenges.

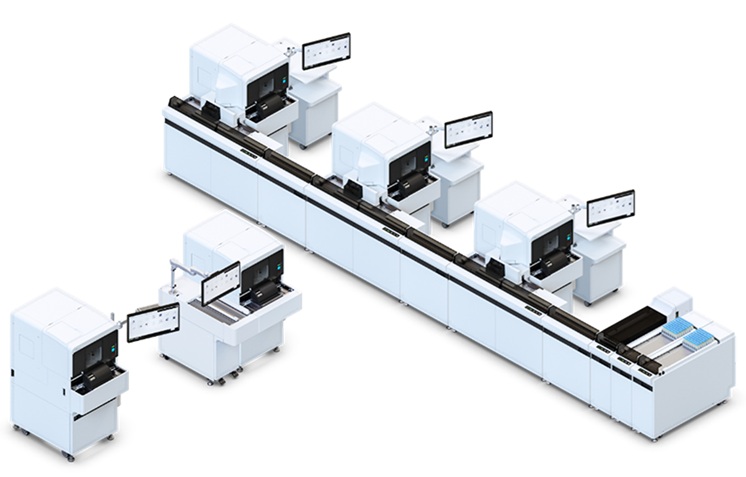
Image: The NovaSeq 6000 Sequencing System (Photo courtesy of Illumina).
A large team of scientists at the University of Texas MD Anderson Cancer Center (Houston, TX, USA) analyzed 154 samples (140 bone marrow mononuclear cells (BMMCs) and 14 peripheral blood mononuclear cells) from 123 patients with acute myeloid leukemia (AML) who had at least one somatic mutation covered by the targeted panel for scDNA-seq. Of the 123 patients, 108 patients were analyzed for the single-timepoint sample collected at pre-treatment (N = 98) or relapsed/refractory timepoint (N = 10). Among 123 patients, 97 were analyzed by scDNA-seq, 23 were analyzed by the simultaneous single-cell DNA and cell surface protein sequencing (scDNA+protein-seq), and three were analyzed by scDNA-seq and scDNA+protein-seq.
The pooled library was sequenced by one of the following sequencing platforms, MiSeq, HiSeq 4000, or NovaSeq 6000 (Illumina, Sand Diego, CA, USA) with 150- or 250-base pair (bp) paired-end multiplexed runs. The team performed droplet digital PCR (ddPCR) using QX200 Droplet Digital System (Bio-Rad Laboratories, Hercules, CA, USA) to confirm the variants that were detected by scDNA-seq, but were not detected by bulk-seq. Simultaneous profiling of DNA mutation and cell-surface immunophenotype (scDNA+protein-seq) was performed using the custom-designed panel kit and 10–15 oligo-conjugated antibodies (Mission Bio, South San Francisco, CA, USA). Immunophenotypes of the bone marrow cells from AML patients were assessed using eight-color flow cytometry on FACSCanto II (BD Biosciences, San Jose, CA, USA).
In all, the scientists sequenced more than 730,000 cells to find 543 somatic mutations in 31 cancer-related genes, 98% of which they orthogonally validated. The most common mutations they detected were in NPM1, followed by ones in DNMT3A and NRAS. They further found that while a number of mutations that were functionally redundant were found in the same patients, the alterations were often found in mutually exclusive clones. This extended to alterations affecting receptor tyrosine kinase (RTK)/Gas GTPase (RAS)/MAP kinase (MAPK) signaling pathway genes as well as IDH1 and IDH2 mutations and TET2 and IDH mutations. This suggested to the scientists that cells either do not need two mutations or that, when they appear together, the mutations are toxic, which could suggest a potential treatment avenue to investigate.
The investigators also analyzed genotype-phenotype correlations among the cells to find, for instance, that cells with NPM1 or IDH mutations expressed lower levels of CD34 and HLA-DR, while cells with a single TP53 mutations had CD34+CD117+ phenotype, but double TP53 mutations had a monocytic immunophenotype.
Koichi Takahashi, MD, PhD, the senior author of the study, said, “This information is also somewhat available from longitudinal bulk sequencing data longitudinally, but I think single-cell data uniquely provides this meticulous view of clone-by-clone dynamics, which is just simply not possible by bulk sequencing.” The study was published on October 21, 2020 in the journal Nature Communications.