Functional Genomic Landscape of AML Analyzed
By LabMedica International staff writers
Posted on 01 Nov 2018
Acute myeloid leukemia is a cancer of the myeloid line of blood cells, characterized by the rapid growth of abnormal cells that build up in the bone marrow and blood and interfere with normal blood cells.Posted on 01 Nov 2018
The implementation of targeted therapies for acute myeloid leukemia (AML) has been challenging because of the complex mutational patterns within and across patients as well as a dearth of pharmacologic agents for most mutational events.

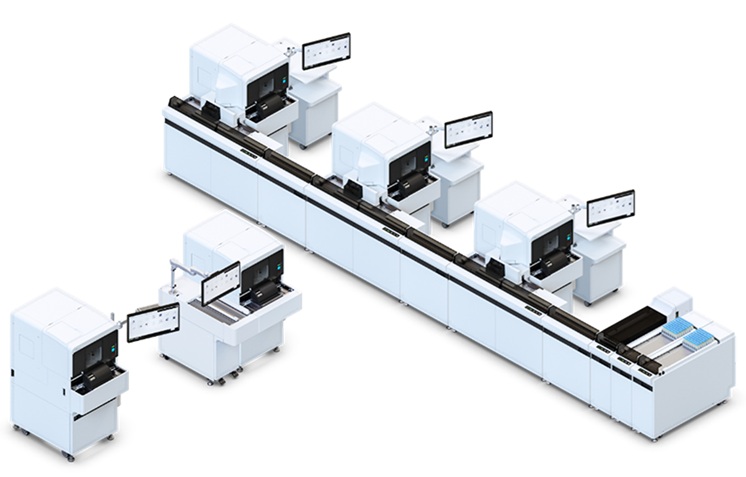
Image: The HiSeq 2500 System is a powerful high-throughput sequencing system (Photo courtesy of Illumina).
Scientists at the Oregon Health & Science University (Portland, OR, USA) used exome sequencing, RNA sequencing, and/or ex vivo drug sensitivity testing to profile up to 672 tumor samples from 562 AML patients. In addition to identifying recurrent mutations not described in the condition previously, they uncovered treatment response patterns that corresponded to specific mutation combinations or gene expression signatures.
As part of a program called Beat AML, the team did paired-end Illumina HiSeq 2500 exome sequencing on 622 of the tumor samples. They also performed RNA sequencing on 451 tumor samples from 411 AML patients, and drug sensitivity testing with 122 drugs on 409 tumor samples from 363 AML patients.
The team uncovered transcriptome profiles associated with frequent tumor mutations or tumor subsets with specific cytogenetic features, as well as gene expression and mutation combinations present when tumors did or did not response to various drugs. Where drug resistance was common in tumors containing mutations in TP53, NRAS, KRAS, IDH1, or the transcriptional regulator-coding gene ASXL1, they noted that IDH2 mutation-positive tumors tended toward drug sensitivity. The team's analysis also highlighted specific drug sensitivities corresponding with targeted treatments such as MAP kinase, PIK3C, mTOR, or JAK inhibitors. These and other data are available to other scientists online through a data viewer called Vizome.
Jeffrey Tyner, PhD, an associate professor and the lead author of the study, said, “"Our dataset can be useful to see if that particular gene mutation corresponds with certain drug sensitivities. We believe this dataset will help physicians solve those specific kinds of questions more easily.” The study was published on October 17, 2018, in the journal Nature.
Related Links:
Oregon Health & Science University