Multiple Myeloma Risk Genes Revealed
By LabMedica International staff writers
Posted on 23 Feb 2018
Two genes have been uncovered with apparent ties to inherited multiple myeloma risk, starting from analyses of several large American families that are frequently affected by the complex blood plasma cell cancer.Posted on 23 Feb 2018
The high-risk pedigree (HRP) design is an established strategy to discover rare, highly penetrant, Mendelian-like causal variants. Its success, however, in complex traits has been modest, largely due to challenges of genetic heterogeneity and complex inheritance models.
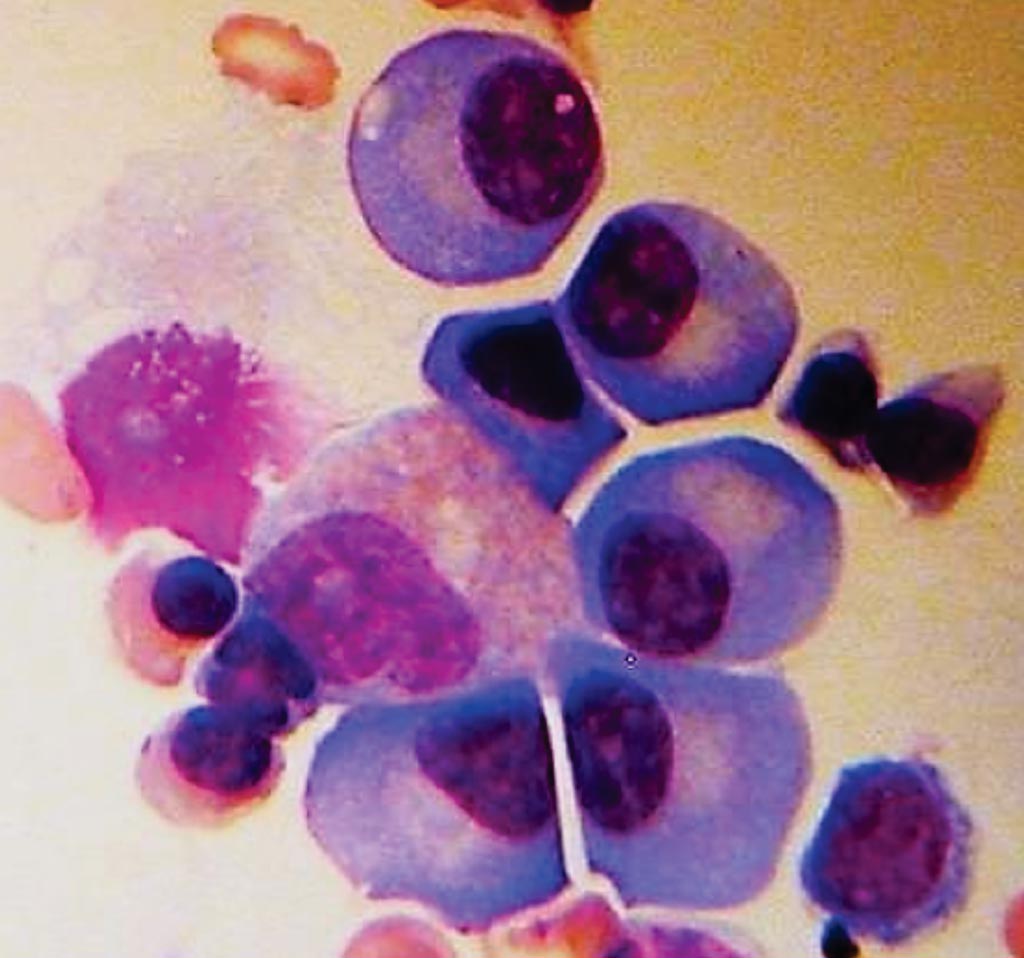
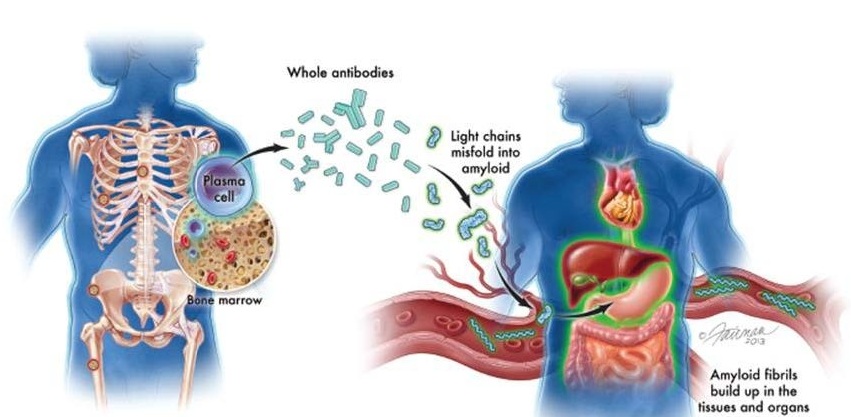
Image: A bone marrow biopsy showing plasma cells from a patient with multiple myeloma (Photo courtesy of the American Society of Clinical Oncology).
An international team of scientists led by those at the University of Utah School of Medicine (Salt Lake City, UT, USA) used an updated version of a gene mapping method known as shared genomic segment (SGS) to narrow in on commonly altered regions in the genomes of multiple myeloma-prone families or "high-risk pedigrees" (HRP) from Utah. By folding in exome sequence data for nearly 1,000 myeloma-free individuals and more than 1,000 individuals affected by multiple myeloma or a precursor condition known as "monoclonal gammopathy of undetermined significance " (MGUS), the team went on to uncover suspicious changes in two genes, Ubiquitin Specific Peptidase 45 (USP45) and AT-Rich Interaction Domain 1A (ARID1A), that appeared to coincide with multiple myeloma risk.
The team used OmniExpress high-density SNP arrays and sequenced individuals from 11 multiple myeloma HRPs, identified through the Utah Cancer Registry, and applied SGS to find shared regions of the genome on chromosomes 6 and 1 with potential ties to multiple myeloma. The team folded in exome sequence data for 28 individuals from the initial 11 pedigrees, along with 126 exomes from a broader set of 44 multiple myeloma pedigrees from the USA, Canada, and France, to identify rare, potentially deleterious mutations in two genes falling the regions of interest: USP45 and ARID1A.
In an even wider exome sequence collection that encompassed 186 early onset multiple myeloma or familial MGUS cases, 733 cases of sporadic multiple myeloma profiled previously, and 964 unaffected controls, the investigators uncovered additional mutations affecting these genes or other genes falling in the same DNA repair or chromatin remodeling complex pathways as USP45 and ARID1A.
The team noted that “Our myeloma findings demonstrate our high-risk pedigree method can identify genetic regions of interest in large, high-risk pedigrees that are also relevant to smaller nuclear families and overall disease risk. We have developed a strategy for gene mapping in complex traits that accounts for heterogeneity within HRPs and formally corrects for multiple testing to allow for statistically rigorous discovery.” The study was published on February 1, 2018, in the journal Public Library of Science Genetics.
Related Links:
University of Utah School of Medicine