Chromosome Interaction Profiling Reveals Cancer Risk Genes
|
By LabMedica International staff writers Posted on 29 Mar 2018 |

Image: The HiSeq2000 instrument used for whole-genome sequencing targeted resequencing, gene expression level detection, DNA methylation profiling, de novo sequencing, metagenomic studies, and ChiP-seq (Photo courtesy of Illumina).
Breast cancer is a heterogeneous disease with two main subtypes defined by the presence (ER+) or absence (ER−) of the estrogen receptor. Approximately 80% of newly diagnosed breast cancers are ER+ although this proportion varies with age at diagnosis and ethnicity.
Genome-wide association studies (GWAS) coupled with large-scale replication and fine-mapping studies have led to the identification of approximately 100 breast cancer risk loci. Potential target and survival-related genes in breast cancer have been identified using chromatin interaction profiles to map parts of the genome previously implicated in risk of the disease.
A team of scientists led by those at the Institute of Cancer Research (London, UK) used a long-range interaction profiling technique called Capture Hi-C (CHi-C) to investigate dozens of regions previously implicated in breast cancer risk. Based on CHi-C data for four breast cancer cell lines with or without enhanced estrogen receptor (ER) activity, a cancer-free breast epithelial cell line, and a cell line from another tissue type, they focused in on 110 potential target genes at 33 of the risk loci.
By adding in related risk single nucleotide polymorphism (SNP) data, RNA sequence profiles, and somatic mutation data reported previously, the team looked at overlap between these proposed targets, expression quantitative trait loci (eQTL), and genes prone to mutation in breast cancer tumors. With the help of gene expression and outcome data from the Metabric breast cancer cohort, meanwhile, the authors highlighted 32 genes with ties to breast cancer survival time. After using HiSeq2000 instruments to sequence target-enriched Hi-C libraries, the team tallied the CHi-C interaction peaks at each risk locus. A dozen risk loci lacked discernable interaction peaks, leaving interaction peaks at 51 loci for further consideration within and across the six cell lines.
The team uncovered potential gene targets at 33 of the breast cancer-associated sites, spanning 94 protein-coding and 16 non-coding RNA gene targets neighboring the risk loci or falling more far afield. They subsequently considered the target genes alongside eQTLs (informed by RNA sequence data from the Cancer Genome Atlas) and somatic mutation profiles identified in hundreds of breast cancer genome sequences, which supported the notion that CHi-C can help unearth authentic target genes.
The authors concluded that a high-throughput CHi-C analysis can contribute to on-going efforts to functionally annotate GWAS risk loci and that CHi-C target genes that are supported by additional data sources are strong candidates for in-depth functional follow-up studies. The study was published on March 12, 2018, in the journal Nature Communications.
Related Links:
Institute of Cancer Research
Genome-wide association studies (GWAS) coupled with large-scale replication and fine-mapping studies have led to the identification of approximately 100 breast cancer risk loci. Potential target and survival-related genes in breast cancer have been identified using chromatin interaction profiles to map parts of the genome previously implicated in risk of the disease.
A team of scientists led by those at the Institute of Cancer Research (London, UK) used a long-range interaction profiling technique called Capture Hi-C (CHi-C) to investigate dozens of regions previously implicated in breast cancer risk. Based on CHi-C data for four breast cancer cell lines with or without enhanced estrogen receptor (ER) activity, a cancer-free breast epithelial cell line, and a cell line from another tissue type, they focused in on 110 potential target genes at 33 of the risk loci.
By adding in related risk single nucleotide polymorphism (SNP) data, RNA sequence profiles, and somatic mutation data reported previously, the team looked at overlap between these proposed targets, expression quantitative trait loci (eQTL), and genes prone to mutation in breast cancer tumors. With the help of gene expression and outcome data from the Metabric breast cancer cohort, meanwhile, the authors highlighted 32 genes with ties to breast cancer survival time. After using HiSeq2000 instruments to sequence target-enriched Hi-C libraries, the team tallied the CHi-C interaction peaks at each risk locus. A dozen risk loci lacked discernable interaction peaks, leaving interaction peaks at 51 loci for further consideration within and across the six cell lines.
The team uncovered potential gene targets at 33 of the breast cancer-associated sites, spanning 94 protein-coding and 16 non-coding RNA gene targets neighboring the risk loci or falling more far afield. They subsequently considered the target genes alongside eQTLs (informed by RNA sequence data from the Cancer Genome Atlas) and somatic mutation profiles identified in hundreds of breast cancer genome sequences, which supported the notion that CHi-C can help unearth authentic target genes.
The authors concluded that a high-throughput CHi-C analysis can contribute to on-going efforts to functionally annotate GWAS risk loci and that CHi-C target genes that are supported by additional data sources are strong candidates for in-depth functional follow-up studies. The study was published on March 12, 2018, in the journal Nature Communications.
Related Links:
Institute of Cancer Research
Latest Molecular Diagnostics News
- Blood Test Accurately Predicts Lung Cancer Risk and Reduces Need for Scans
- Unique Autoantibody Signature to Help Diagnose Multiple Sclerosis Years before Symptom Onset
- Blood Test Could Detect HPV-Associated Cancers 10 Years before Clinical Diagnosis
- Low-Cost Point-Of-Care Diagnostic to Expand Access to STI Testing
- 18-Gene Urine Test for Prostate Cancer to Help Avoid Unnecessary Biopsies
- Urine-Based Test Detects Head and Neck Cancer
- Blood-Based Test Detects and Monitors Aggressive Small Cell Lung Cancer
- Blood-Based Machine Learning Assay Noninvasively Detects Ovarian Cancer
- Simple PCR Assay Accurately Differentiates Between Small Cell Lung Cancer Subtypes
- Revolutionary T-Cell Analysis Approach Enables Cancer Early Detection
- Single Genetic Test to Accelerate Diagnoses for Rare Developmental Disorders
- Upgraded Syndromic Testing Analyzer Enables Remote Test Results Access
- Respiratory and Throat Infection PCR Test Detects Multiple Pathogens with Overlapping Symptoms
- Blood Circulating Nucleic Acid Enrichment Technique Enables Non-Invasive Liver Cancer Diagnosis
- First FDA-Approved Molecular Test to Screen Blood Donors for Malaria Could Improve Patient Safety
- Fluid Biomarker Test Detects Neurodegenerative Diseases Before Symptoms Appear
Channels
Clinical Chemistry
view channel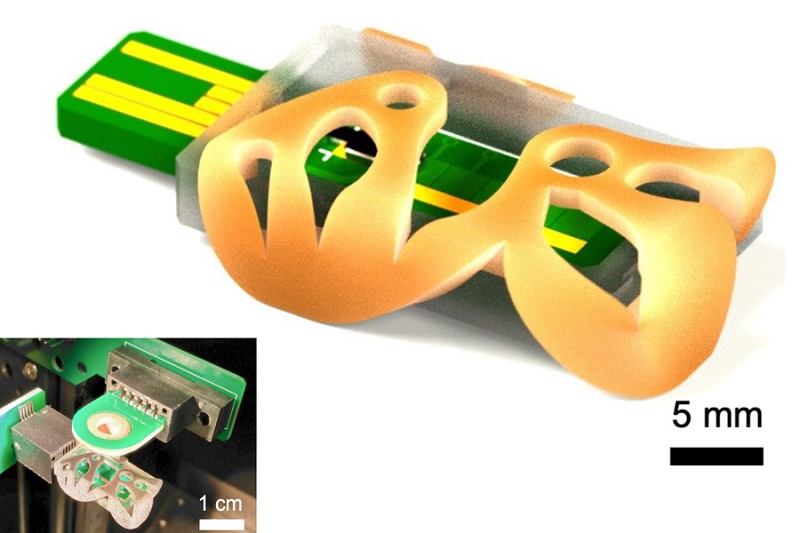
3D Printed Point-Of-Care Mass Spectrometer Outperforms State-Of-The-Art Models
Mass spectrometry is a precise technique for identifying the chemical components of a sample and has significant potential for monitoring chronic illness health states, such as measuring hormone levels... Read more.jpg)
POC Biomedical Test Spins Water Droplet Using Sound Waves for Cancer Detection
Exosomes, tiny cellular bioparticles carrying a specific set of proteins, lipids, and genetic materials, play a crucial role in cell communication and hold promise for non-invasive diagnostics.... Read more
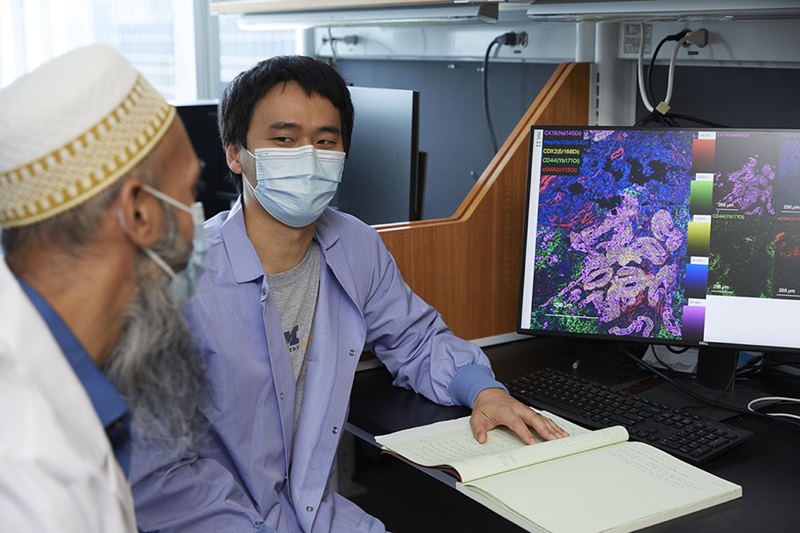
Highly Reliable Cell-Based Assay Enables Accurate Diagnosis of Endocrine Diseases
The conventional methods for measuring free cortisol, the body's stress hormone, from blood or saliva are quite demanding and require sample processing. The most common method, therefore, involves collecting... Read moreHematology
view channel
Next Generation Instrument Screens for Hemoglobin Disorders in Newborns
Hemoglobinopathies, the most widespread inherited conditions globally, affect about 7% of the population as carriers, with 2.7% of newborns being born with these conditions. The spectrum of clinical manifestations... Read more
First 4-in-1 Nucleic Acid Test for Arbovirus Screening to Reduce Risk of Transfusion-Transmitted Infections
Arboviruses represent an emerging global health threat, exacerbated by climate change and increased international travel that is facilitating their spread across new regions. Chikungunya, dengue, West... Read more
POC Finger-Prick Blood Test Determines Risk of Neutropenic Sepsis in Patients Undergoing Chemotherapy
Neutropenia, a decrease in neutrophils (a type of white blood cell crucial for fighting infections), is a frequent side effect of certain cancer treatments. This condition elevates the risk of infections,... Read more
First Affordable and Rapid Test for Beta Thalassemia Demonstrates 99% Diagnostic Accuracy
Hemoglobin disorders rank as some of the most prevalent monogenic diseases globally. Among various hemoglobin disorders, beta thalassemia, a hereditary blood disorder, affects about 1.5% of the world's... Read moreImmunology
view channel
Diagnostic Blood Test for Cellular Rejection after Organ Transplant Could Replace Surgical Biopsies
Transplanted organs constantly face the risk of being rejected by the recipient's immune system which differentiates self from non-self using T cells and B cells. T cells are commonly associated with acute... Read more
AI Tool Precisely Matches Cancer Drugs to Patients Using Information from Each Tumor Cell
Current strategies for matching cancer patients with specific treatments often depend on bulk sequencing of tumor DNA and RNA, which provides an average profile from all cells within a tumor sample.... Read more
Genetic Testing Combined With Personalized Drug Screening On Tumor Samples to Revolutionize Cancer Treatment
Cancer treatment typically adheres to a standard of care—established, statistically validated regimens that are effective for the majority of patients. However, the disease’s inherent variability means... Read moreMicrobiology
view channel
New CE-Marked Hepatitis Assays to Help Diagnose Infections Earlier
According to the World Health Organization (WHO), an estimated 354 million individuals globally are afflicted with chronic hepatitis B or C. These viruses are the leading causes of liver cirrhosis, liver... Read more
1 Hour, Direct-From-Blood Multiplex PCR Test Identifies 95% of Sepsis-Causing Pathogens
Sepsis contributes to one in every three hospital deaths in the US, and globally, septic shock carries a mortality rate of 30-40%. Diagnosing sepsis early is challenging due to its non-specific symptoms... Read morePathology
view channelAI-Powered Digital Imaging System to Revolutionize Cancer Diagnosis
The process of biopsy is important for confirming the presence of cancer. In the conventional histopathology technique, tissue is excised, sliced, stained, mounted on slides, and examined under a microscope... Read more
New Mycobacterium Tuberculosis Panel to Support Real-Time Surveillance and Combat Antimicrobial Resistance
Tuberculosis (TB), the leading cause of death from an infectious disease globally, is a contagious bacterial infection that primarily spreads through the coughing of patients with active pulmonary TB.... Read moreTechnology
view channel
New Diagnostic System Achieves PCR Testing Accuracy
While PCR tests are the gold standard of accuracy for virology testing, they come with limitations such as complexity, the need for skilled lab operators, and longer result times. They also require complex... Read more
DNA Biosensor Enables Early Diagnosis of Cervical Cancer
Molybdenum disulfide (MoS2), recognized for its potential to form two-dimensional nanosheets like graphene, is a material that's increasingly catching the eye of the scientific community.... Read more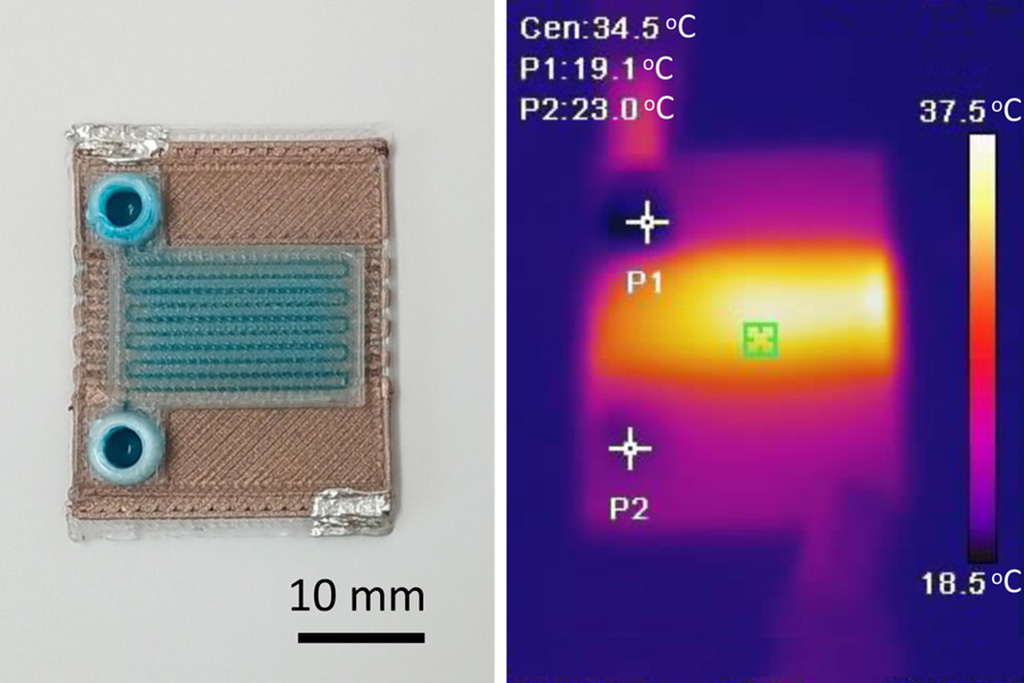
Self-Heating Microfluidic Devices Can Detect Diseases in Tiny Blood or Fluid Samples
Microfluidics, which are miniature devices that control the flow of liquids and facilitate chemical reactions, play a key role in disease detection from small samples of blood or other fluids.... Read more
Breakthrough in Diagnostic Technology Could Make On-The-Spot Testing Widely Accessible
Home testing gained significant importance during the COVID-19 pandemic, yet the availability of rapid tests is limited, and most of them can only drive one liquid across the strip, leading to continued... Read moreIndustry
view channel
ECCMID Congress Name Changes to ESCMID Global
Over the last few years, the European Society of Clinical Microbiology and Infectious Diseases (ESCMID, Basel, Switzerland) has evolved remarkably. The society is now stronger and broader than ever before... Read more
Bosch and Randox Partner to Make Strategic Investment in Vivalytic Analysis Platform
Given the presence of so many diseases, determining whether a patient is presenting the symptoms of a simple cold, the flu, or something as severe as life-threatening meningitis is usually only possible... Read more
Siemens to Close Fast Track Diagnostics Business
Siemens Healthineers (Erlangen, Germany) has announced its intention to close its Fast Track Diagnostics unit, a small collection of polymerase chain reaction (PCR) testing products that is part of the... Read more











.jpg)